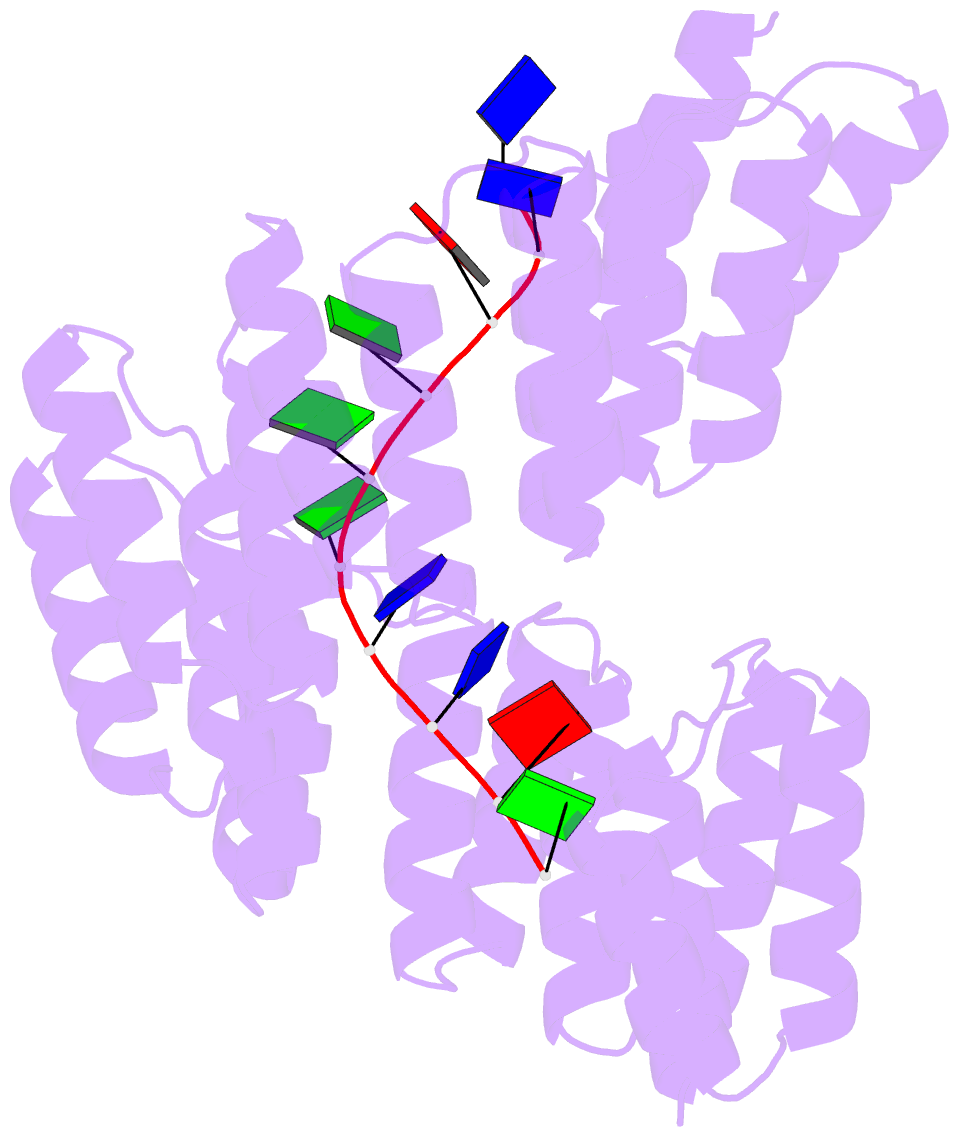
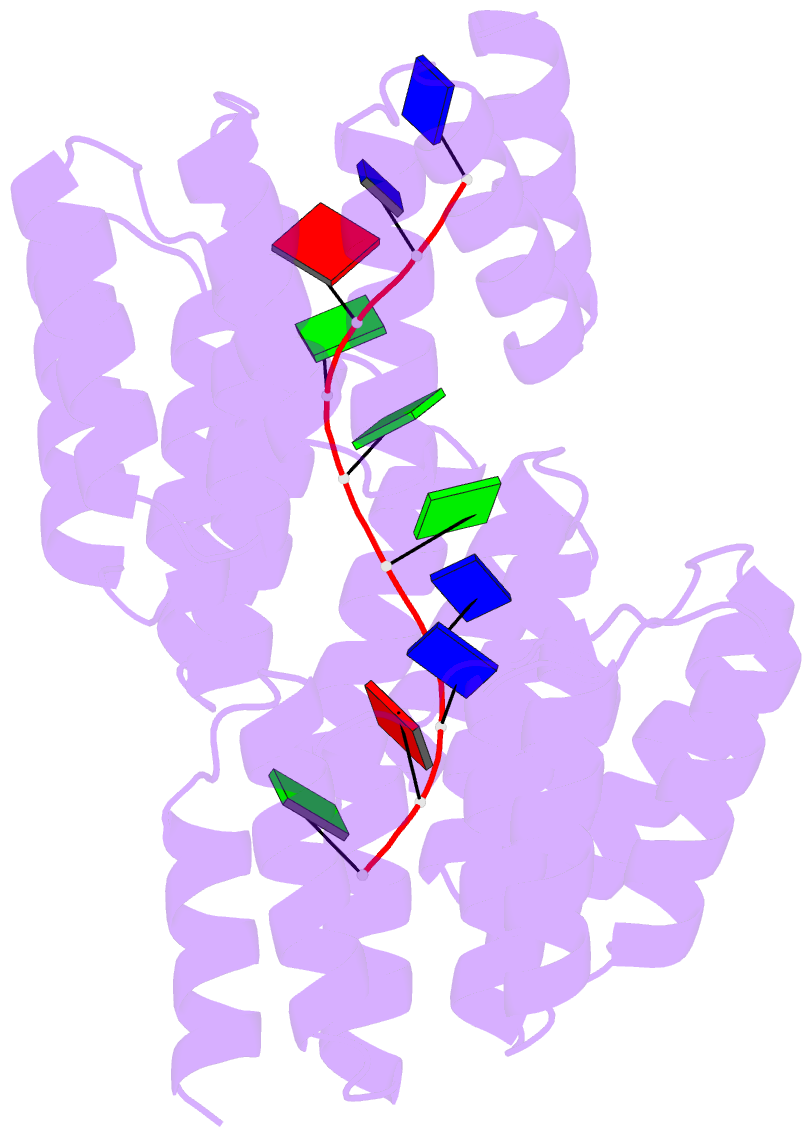
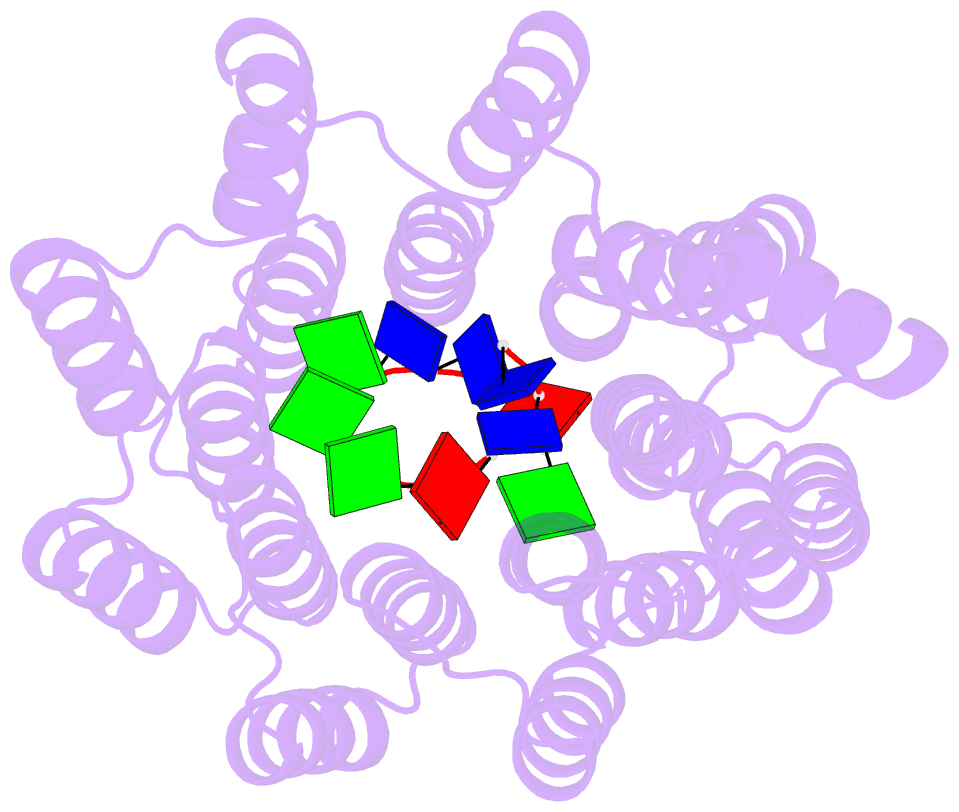
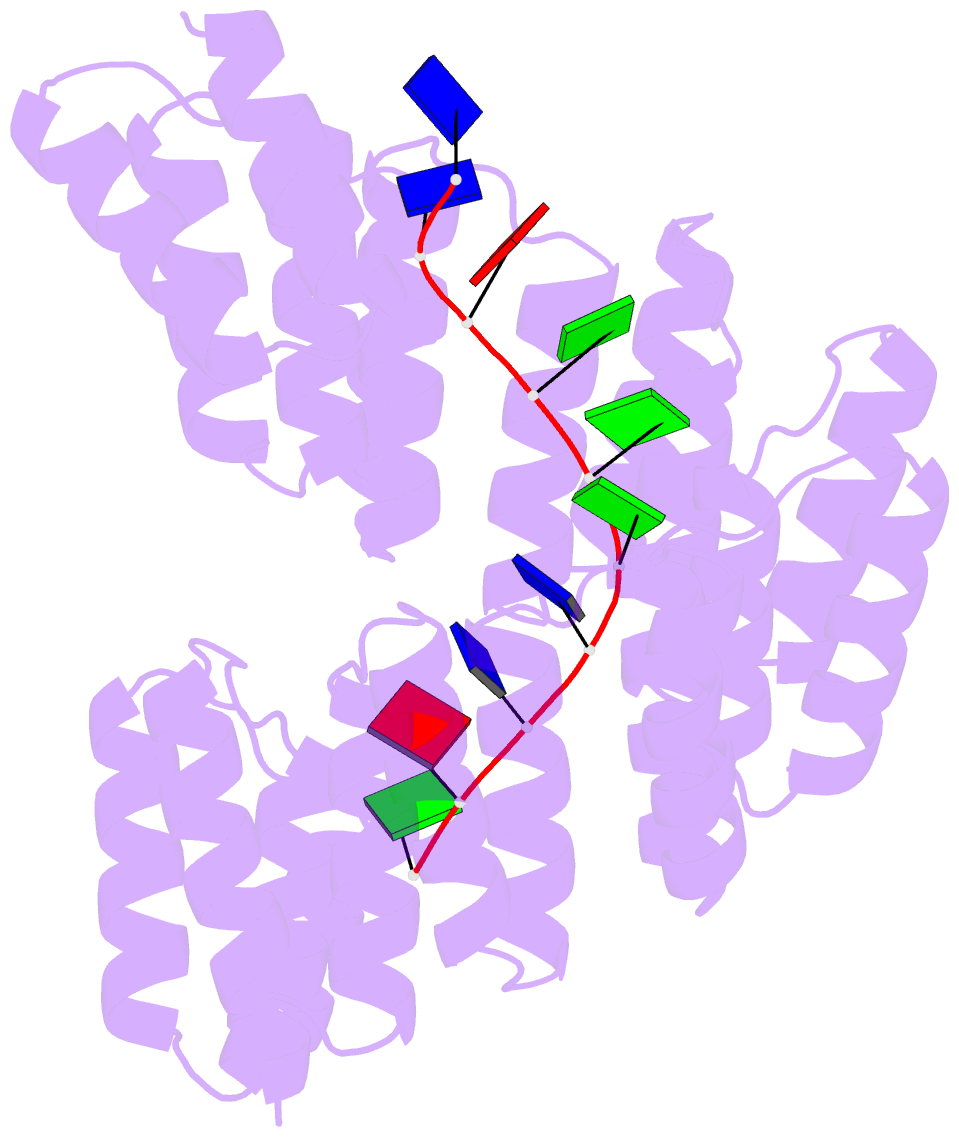
Summary information and primary citation
- PDB-id
- 5orq; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- de novo protein
- Method
- X-ray (1.95 Å)
- Summary
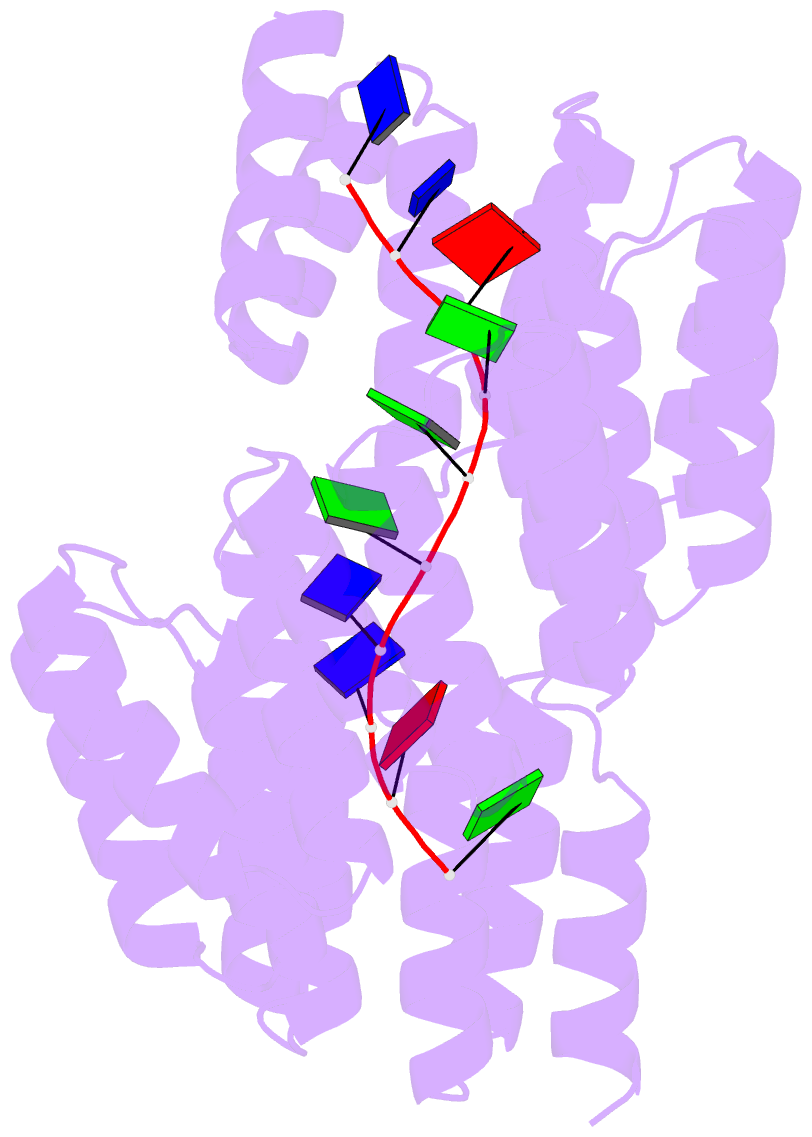
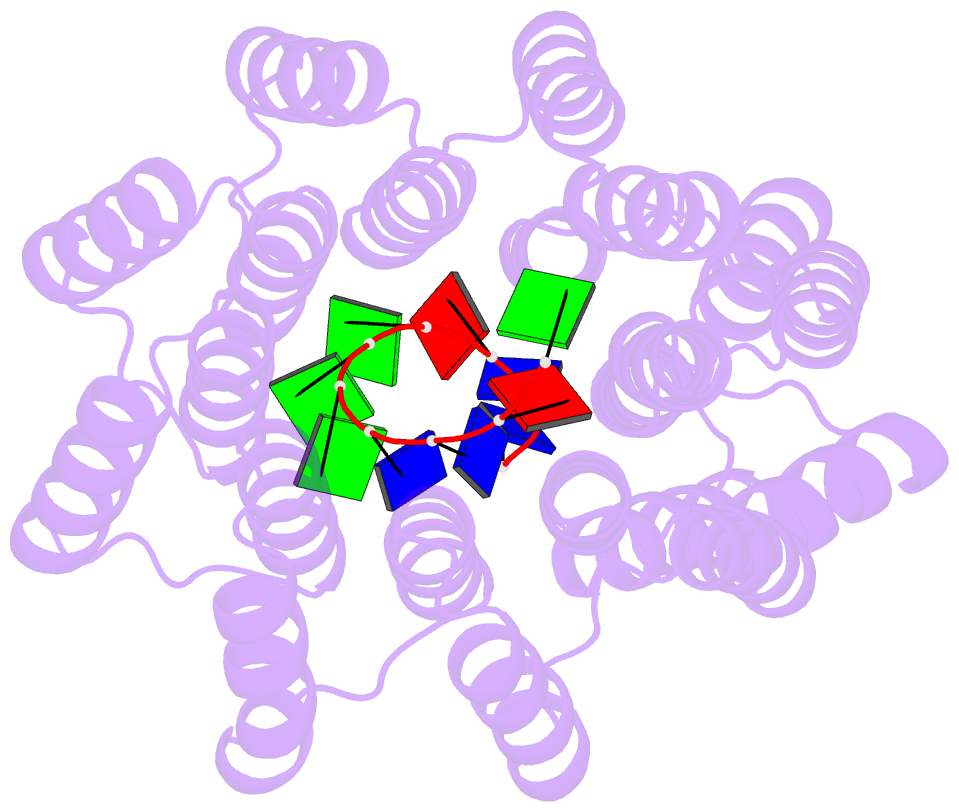
- Crystal structure of designed cppr-telo1 in complex with ssDNA
- Reference
- Spahr H, Chia T, Lingford JP, Siira SJ, Cohen SB, Filipovska A, Rackham O (2018): "Modular ssDNA binding and inhibition of telomerase activity by designer PPR proteins." Nat Commun, 9, 2212. doi: 10.1038/s41467-018-04388-1.
- Abstract
- DNA is typically found as a double helix, however it must be separated into single strands during all phases of DNA metabolism; including transcription, replication, recombination and repair. Although recent breakthroughs have enabled the design of modular RNA- and double-stranded DNA-binding proteins, there are currently no tools available to manipulate single-stranded DNA (ssDNA). Here we show that artificial pentatricopeptide repeat (PPR) proteins can be programmed for sequence-specific ssDNA binding. Interactions occur using the same code and specificity as for RNA binding. We solve the structures of DNA-bound and apo proteins revealing the basis for ssDNA binding and how hydrogen bond rearrangements enable the PPR structure to envelope its ssDNA target. Finally, we show that engineered PPRs can be designed to bind telomeric ssDNA and can block telomerase activity. The modular mode of ssDNA binding by PPR proteins provides tools to target ssDNA and to understand its importance in cells.