Summary information and primary citation
- PDB-id
- 5oxj; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase
- Method
- X-ray (2.0 Å)
- Summary
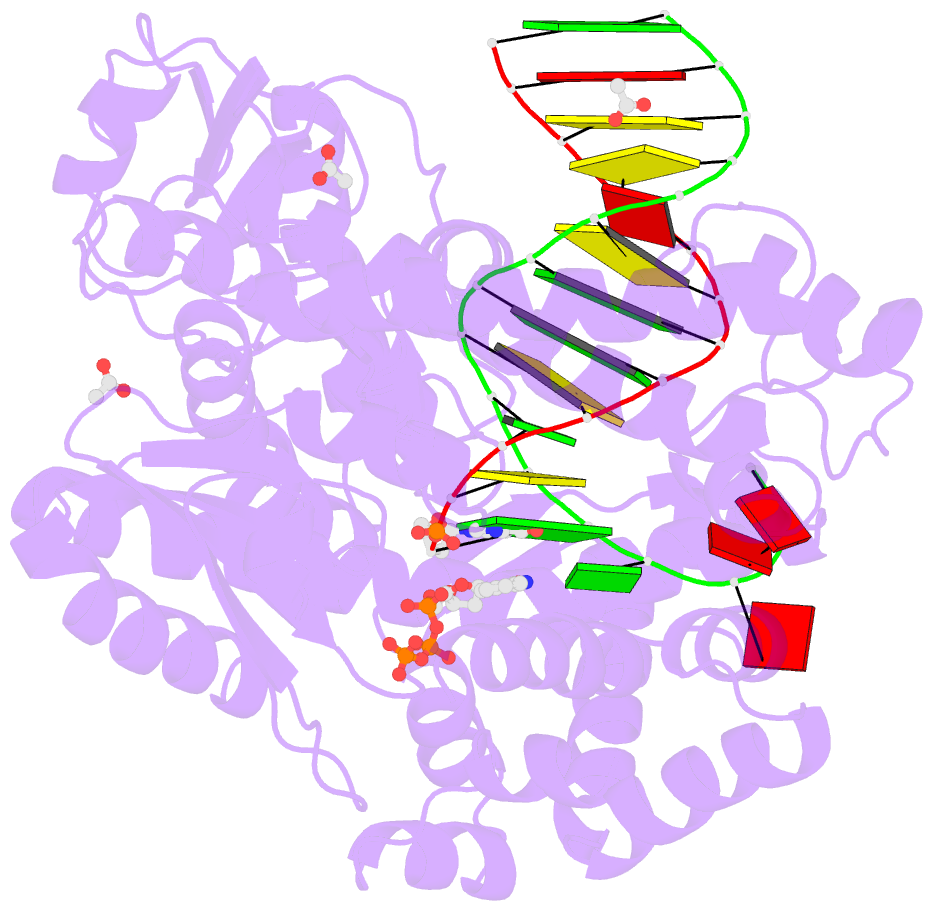
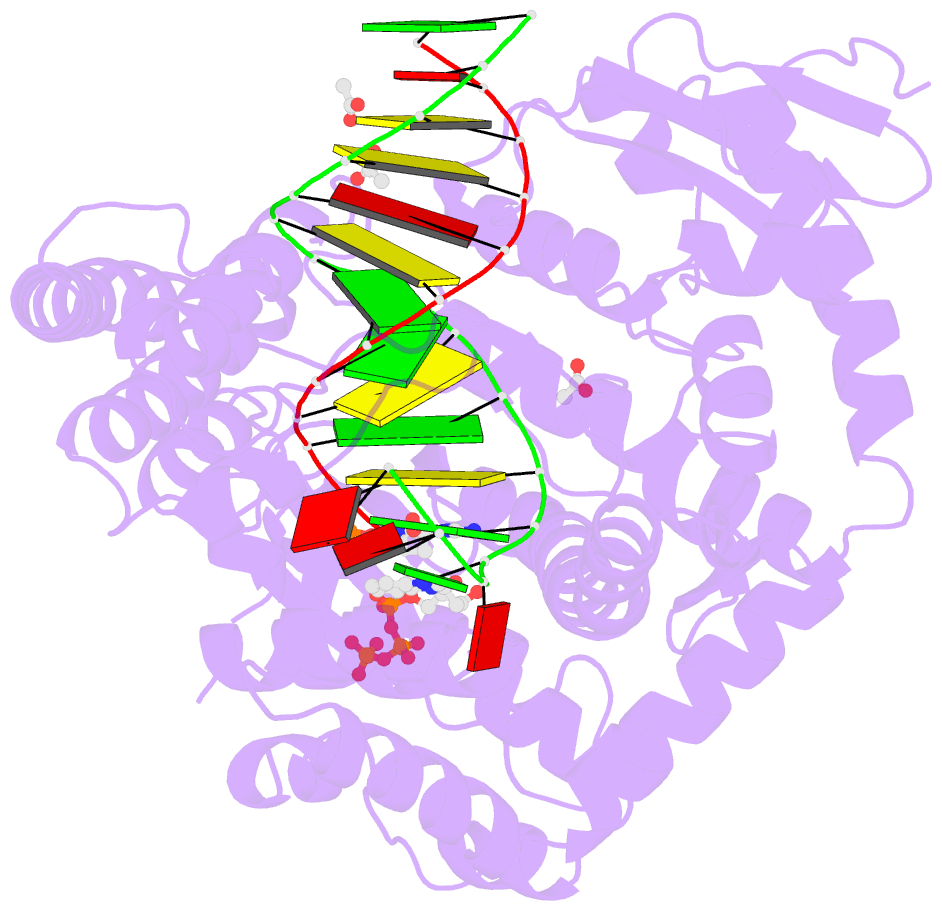
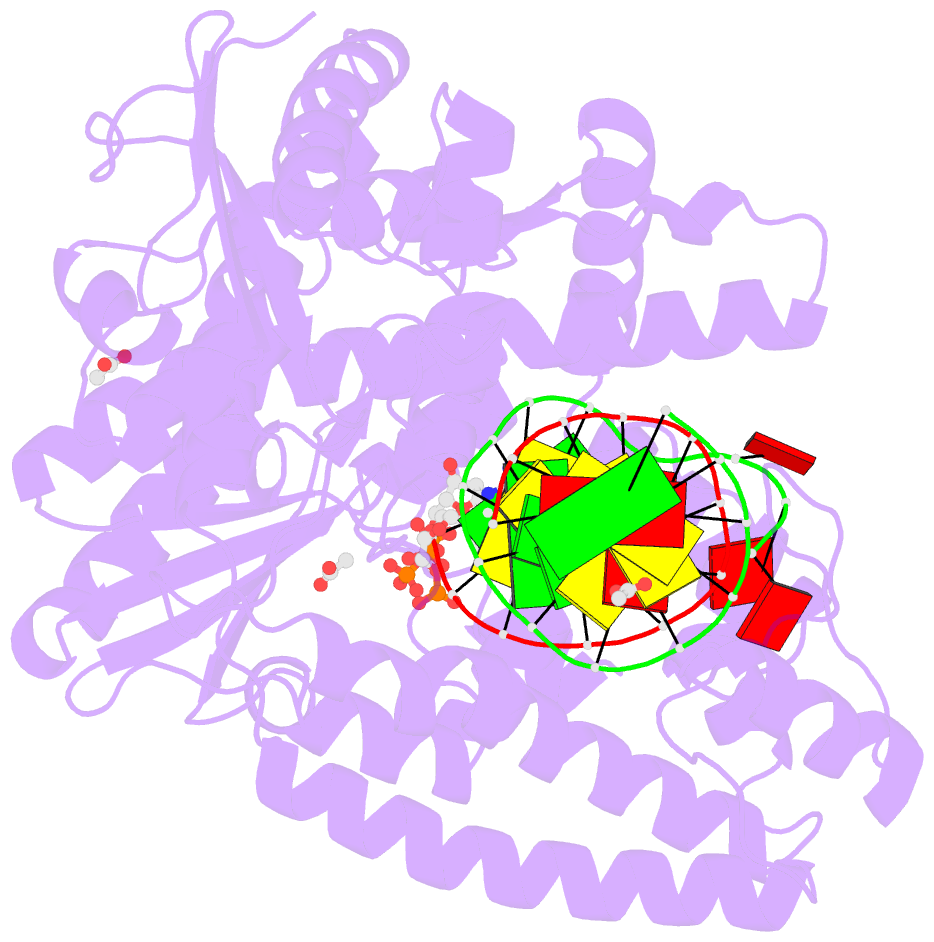
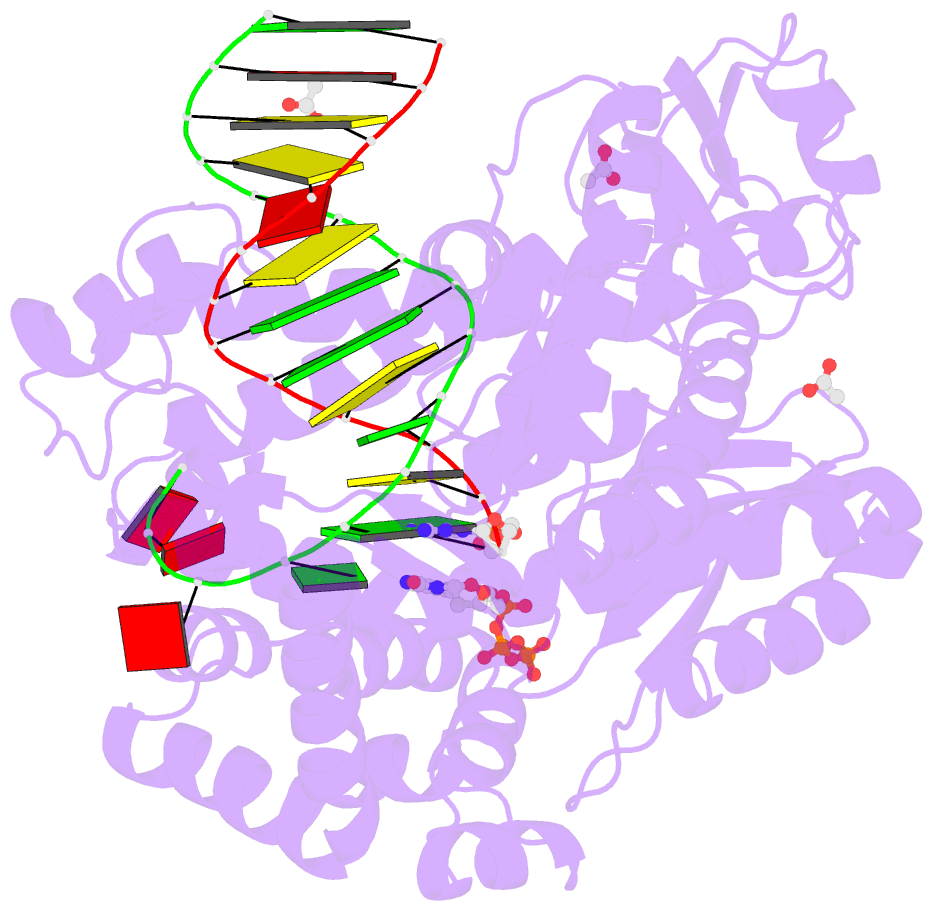
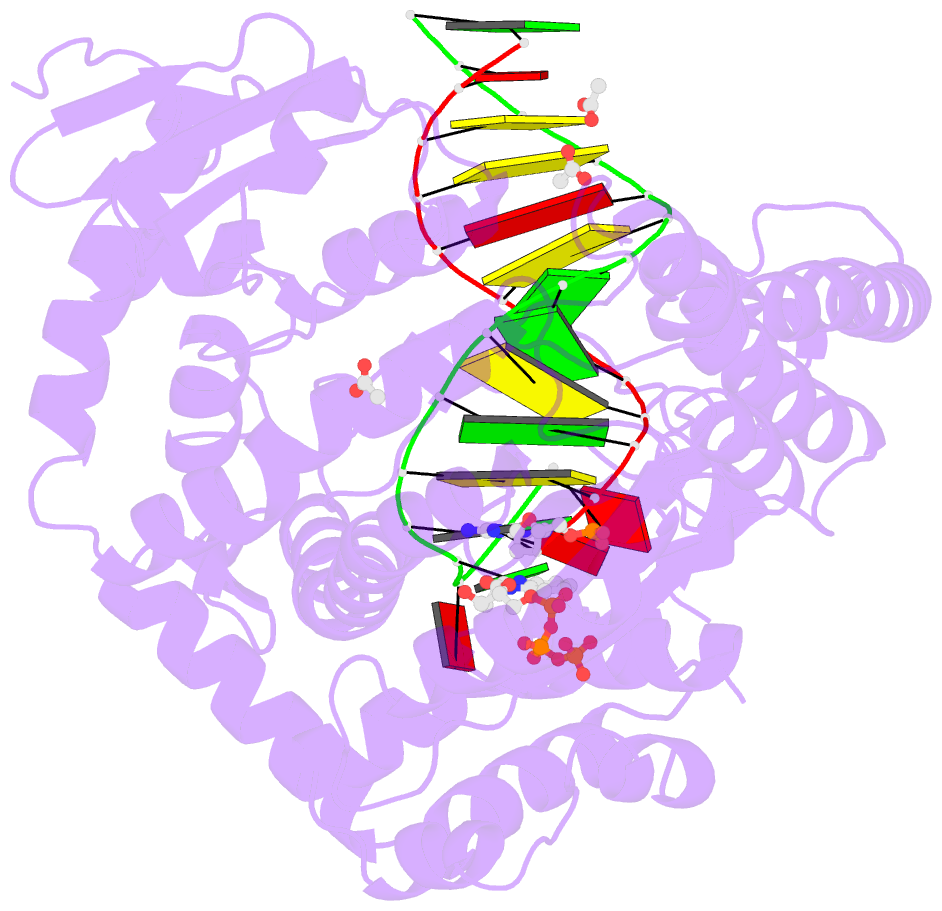
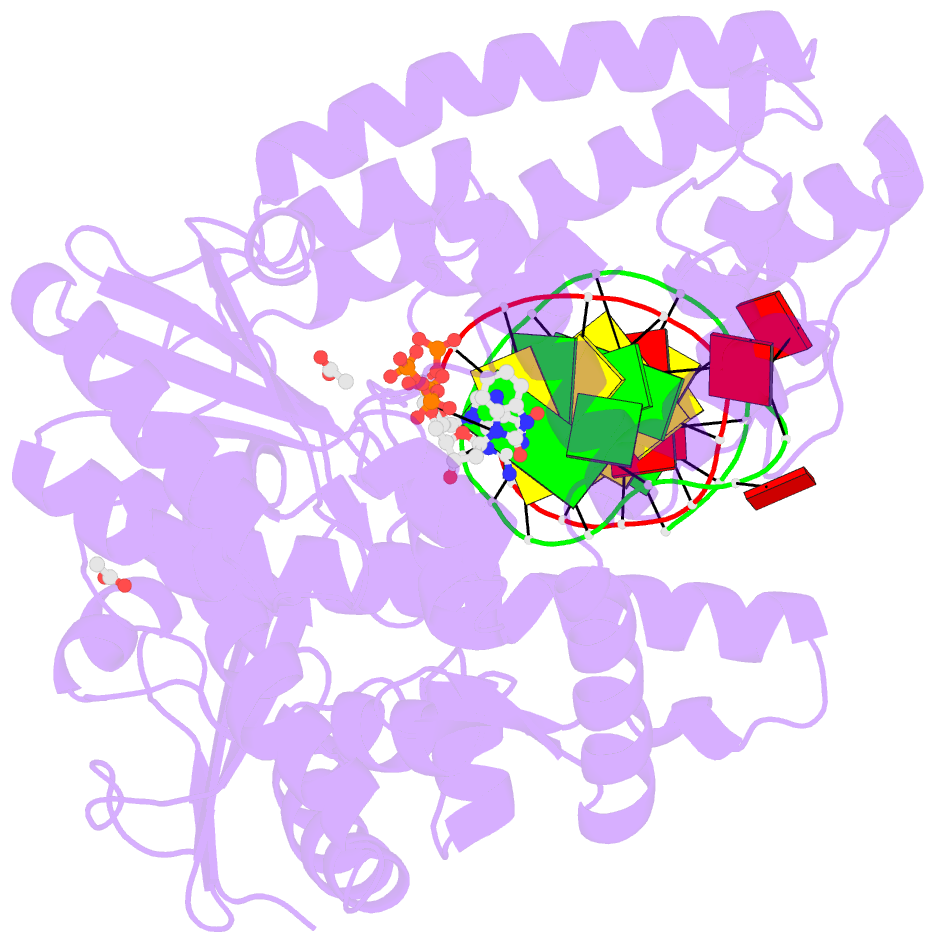
- Crystal structure of klentaq mutant m747k in a closed ternary complex with a o6-meg:benzitp base pair
- Reference
- Betz K, Nilforoushan A, Wyss LA, Diederichs K, Sturla SJ, Marx A (2017): "Structural basis for the selective incorporation of an artificial nucleotide opposite a DNA adduct by a DNA polymerase." Chem. Commun. (Camb.), 53, 12704-12707. doi: 10.1039/c7cc07173f.
- Abstract
- The possibility to sequence cytotoxic O6-alkylG DNA adducts would greatly benefit research. Recently we reported a benzimidazole-derived nucleotide that is selectively incorporated opposite the damaged site by a mutated DNA polymerase. Here we provide the structural basis for this reaction which may spur future developments in DNA damage sequencing.