Summary information and primary citation
- PDB-id
- 5oxv; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- gene regulation
- Method
- X-ray (6.721 Å)
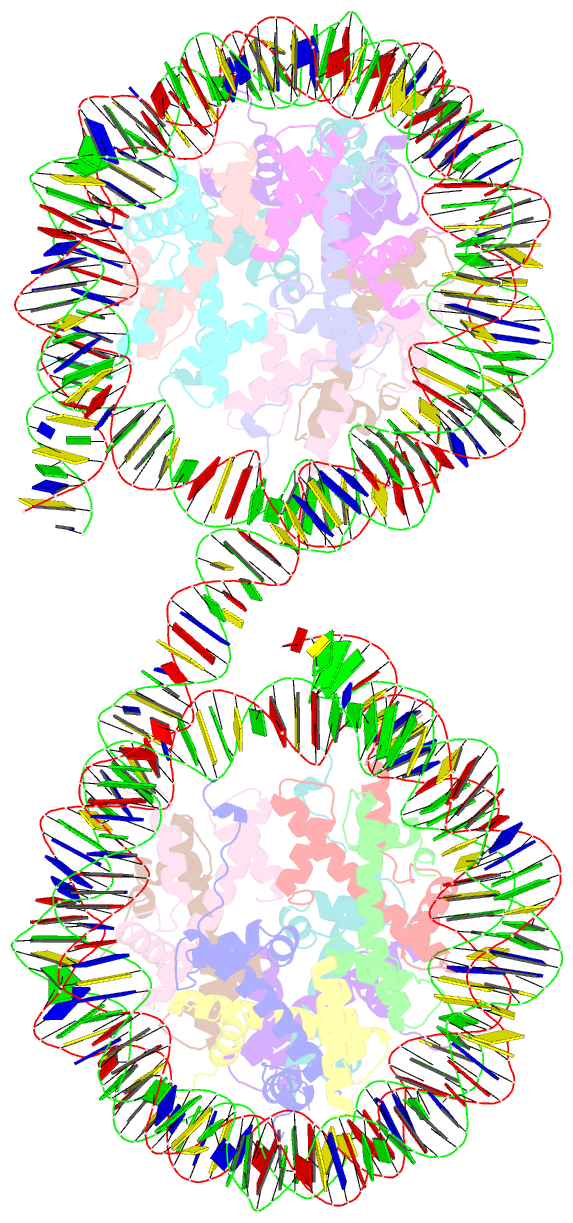
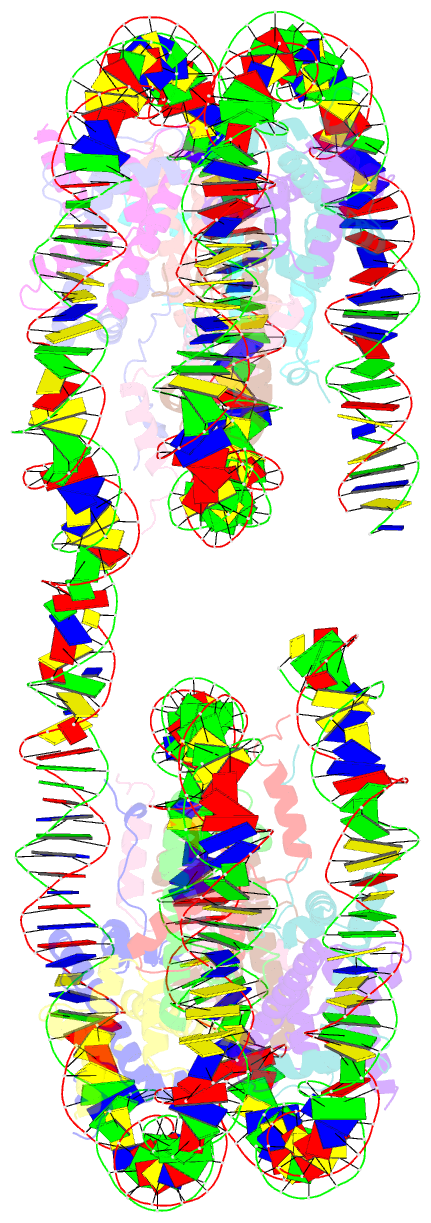
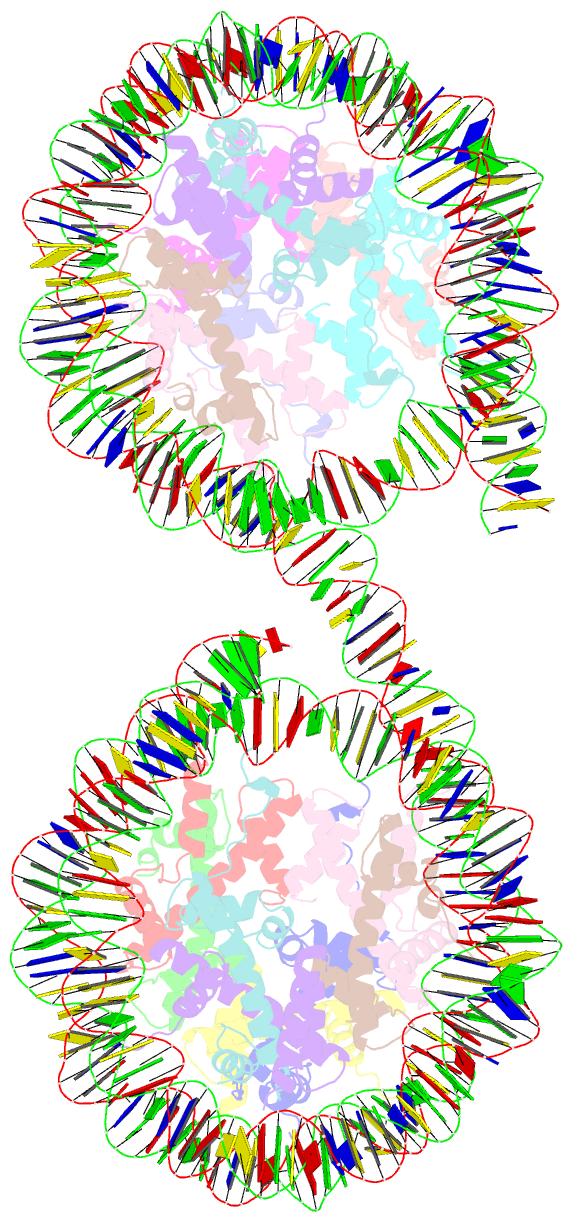
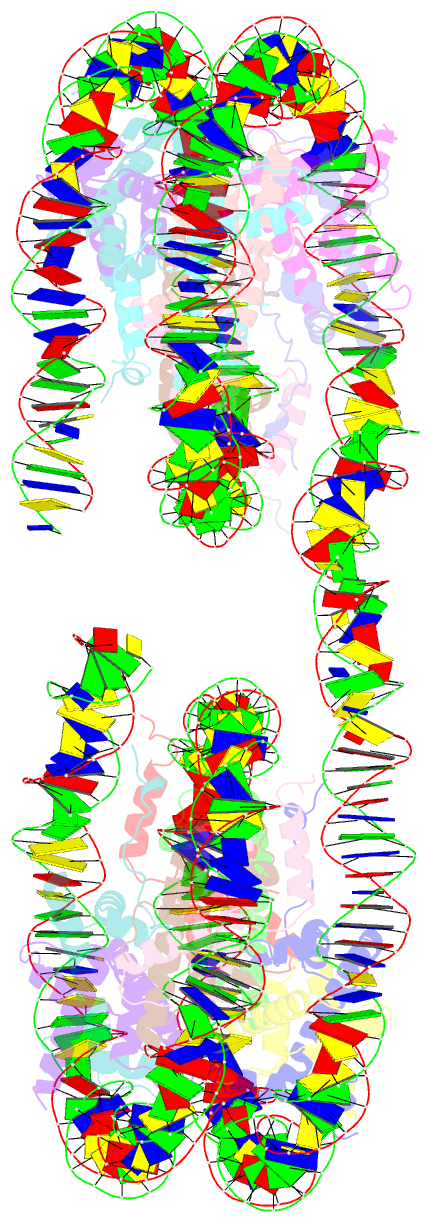
- Summary
- Structure of the 4_601_157 tetranucleosome (c2 form)
- Reference
- Ekundayo B, Richmond TJ, Schalch T (2017): "Capturing Structural Heterogeneity in Chromatin Fibers." J. Mol. Biol., 429, 3031-3042. doi: 10.1016/j.jmb.2017.09.002.
- Abstract
- Chromatin fiber organization is implicated in processes such as transcription, DNA repair and chromosome segregation, but how nucleosomes interact to form higher-order structure remains poorly understood. We solved two crystal structures of tetranucleosomes with approximately 11-bp DNA linker length at 5.8 and 6.7 Å resolution. Minimal intramolecular nucleosome-nucleosome interactions result in a fiber model resembling a flat ribbon that is compatible with a two-start helical architecture, and that exposes histone and DNA surfaces to the environment. The differences in the two structures combined with electron microscopy reveal heterogeneous structural states, and we used site-specific chemical crosslinking to assess the diversity of nucleosome-nucleosome interactions through identification of structure-sensitive crosslink sites that provide a means to characterize fibers in solution. The chromatin fiber architectures observed here provide a basis for understanding heterogeneous chromatin higher-order structures as they occur in a genomic context.