Summary information and primary citation
- PDB-id
- 5sze; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA-binding protein-RNA
- Method
- X-ray (1.5 Å)
- Summary
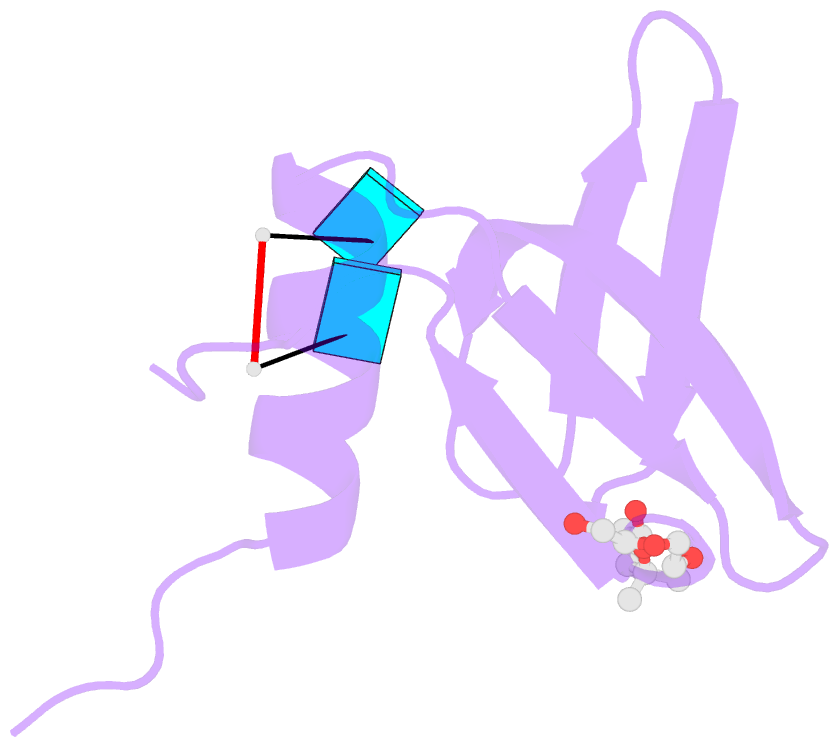
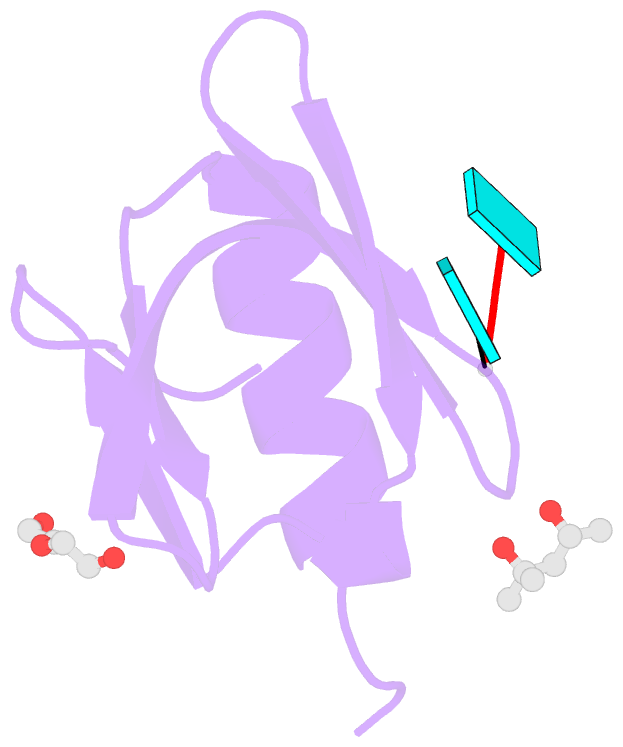
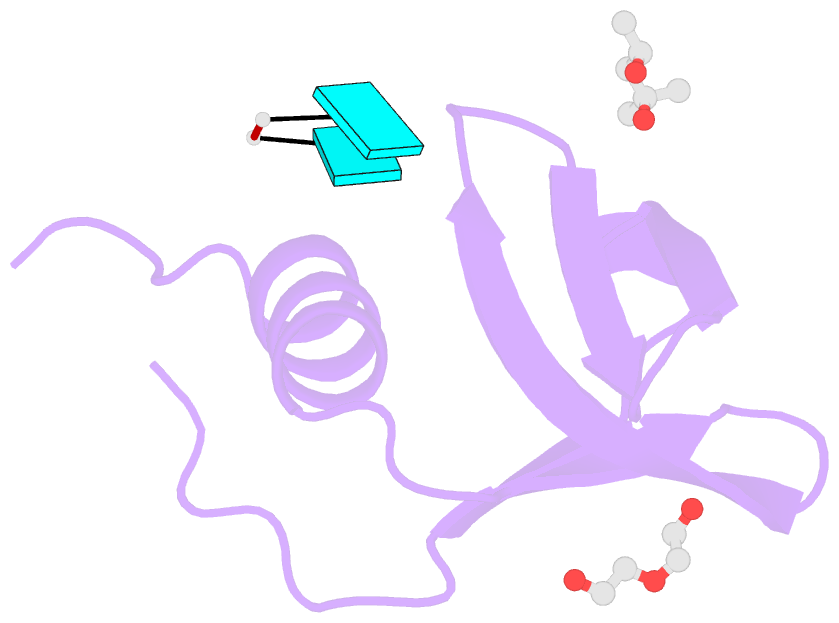
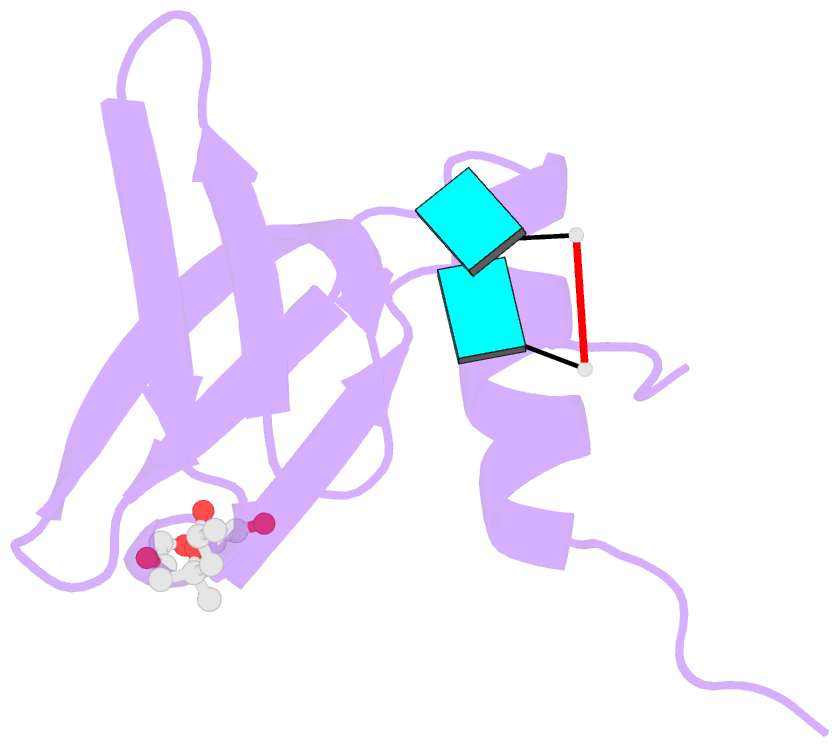
- Crystal structure of aquifex aeolicus hfq-RNA complex at 1.5a
- Reference
- Stanek KA, Patterson-West J, Randolph PS, Mura C (2017): "Crystal structure and RNA-binding properties of an Hfq homolog from the deep-branching Aquificae: conservation of the lateral RNA-binding mode." Acta Crystallogr D Struct Biol, 73, 294-315. doi: 10.1107/S2059798317000031.
- Abstract
- The host factor Hfq, as the bacterial branch of the Sm family, is an RNA-binding protein involved in the post-transcriptional regulation of mRNA expression and turnover. Hfq facilitates pairing between small regulatory RNAs (sRNAs) and their corresponding mRNA targets by binding both RNAs and bringing them into close proximity. Hfq homologs self-assemble into homo-hexameric rings with at least two distinct surfaces that bind RNA. Recently, another binding site, dubbed the `lateral rim', has been implicated in sRNA·mRNA annealing; the RNA-binding properties of this site appear to be rather subtle, and its degree of evolutionary conservation is unknown. An Hfq homolog has been identified in the phylogenetically deep-branching thermophile Aquifex aeolicus (Aae), but little is known about the structure and function of Hfq from basal bacterial lineages such as the Aquificae. Therefore, Aae Hfq was cloned, overexpressed, purified, crystallized and biochemically characterized. Structures of Aae Hfq were determined in space groups P1 and P6, both to 1.5 Å resolution, and nanomolar-scale binding affinities for uridine- and adenosine-rich RNAs were discovered. Co-crystallization with U6 RNA reveals that the outer rim of the Aae Hfq hexamer features a well defined binding pocket that is selective for uracil. This Aae Hfq structure, combined with biochemical and biophysical characterization of the homolog, reveals deep evolutionary conservation of the lateral RNA-binding mode, and lays a foundation for further studies of Hfq-associated RNA biology in ancient bacterial phyla.