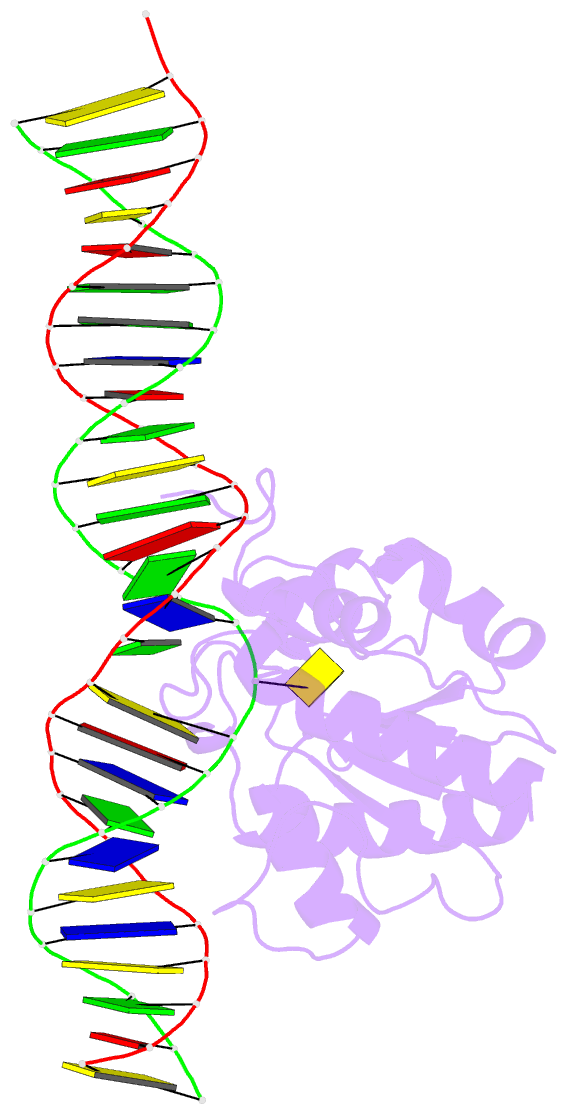
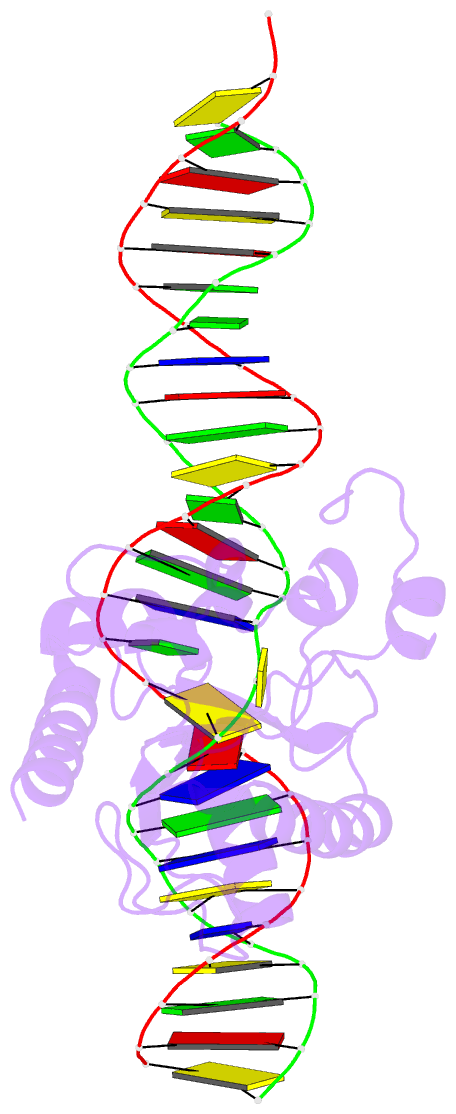
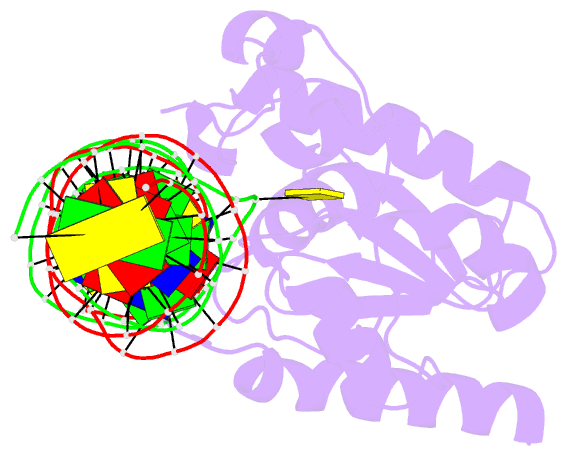
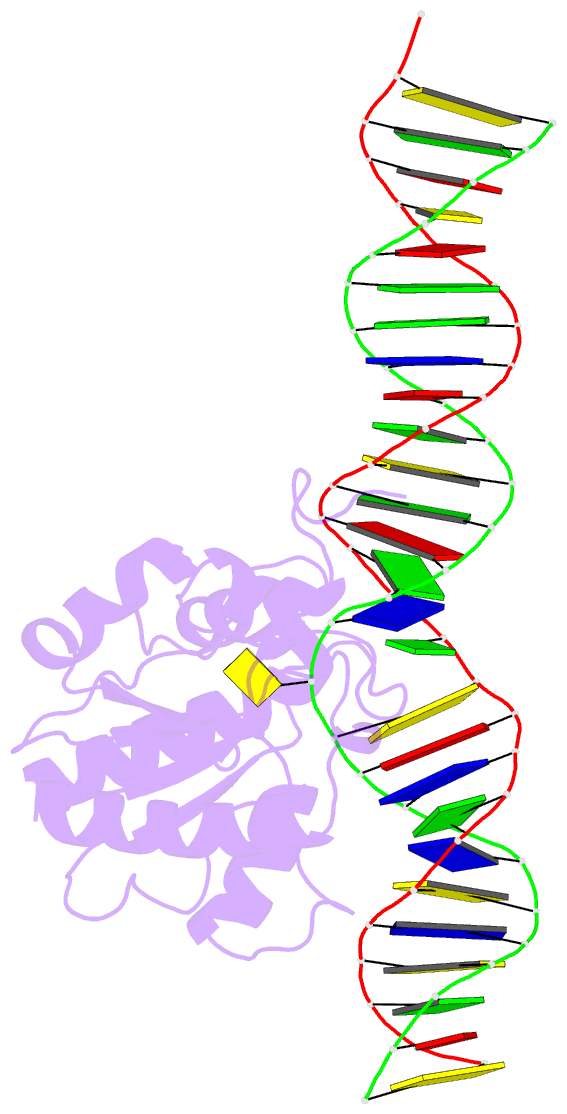
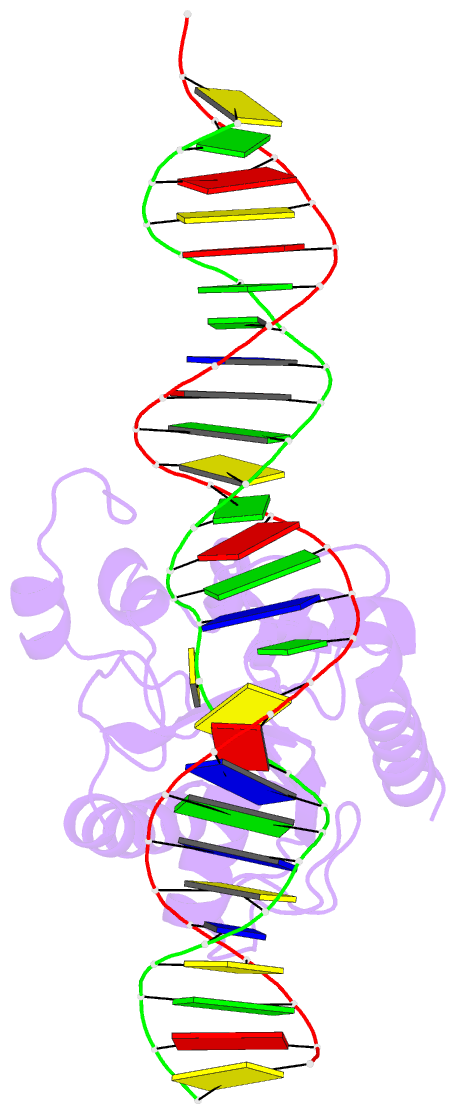
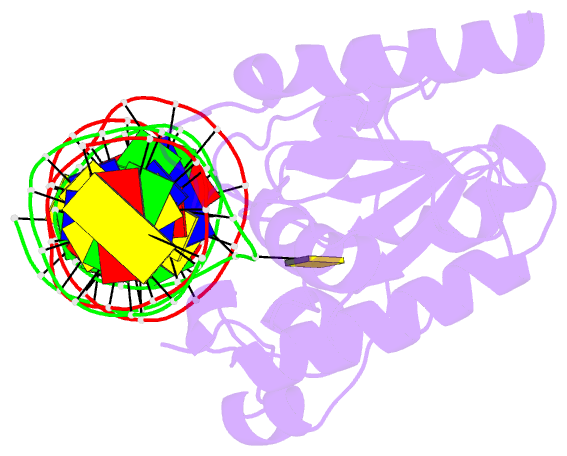
Summary information and primary citation
- PDB-id
- 5t2w; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (2.2 Å)
- Summary
- Structure of thymine DNA glycosylase bound to substrate analog 2'-f-5-formyl-dc
- Reference
- Pidugu LS, Flowers JW, Coey CT, Pozharski E, Greenberg MM, Drohat AC (2016): "Structural Basis for Excision of 5-Formylcytosine by Thymine DNA Glycosylase." Biochemistry, 55, 6205-6208. doi: 10.1021/acs.biochem.6b00982.
- Abstract
- Thymine DNA glycosylase (TDG) is a base excision repair enzyme with key functions in epigenetic regulation. Performing a critical step in a pathway for active DNA demethylation, TDG removes 5-formylcytosine and 5-carboxylcytosine, oxidized derivatives of 5-methylcytosine that are generated by TET (ten-eleven translocation) enzymes. We determined a crystal structure of TDG bound to DNA with a noncleavable (2'-fluoroarabino) analogue of 5-formyldeoxycytidine flipped into its active site, revealing how it recognizes and hydrolytically excises fC. Together with previous structural and biochemical findings, the results illustrate how TDG employs an adaptable active site to excise a broad variety of nucleobases from DNA.