Summary information and primary citation
- PDB-id
- 5t8y; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- oxidoreductase-RNA
- Method
- X-ray (2.653 Å)
- Summary
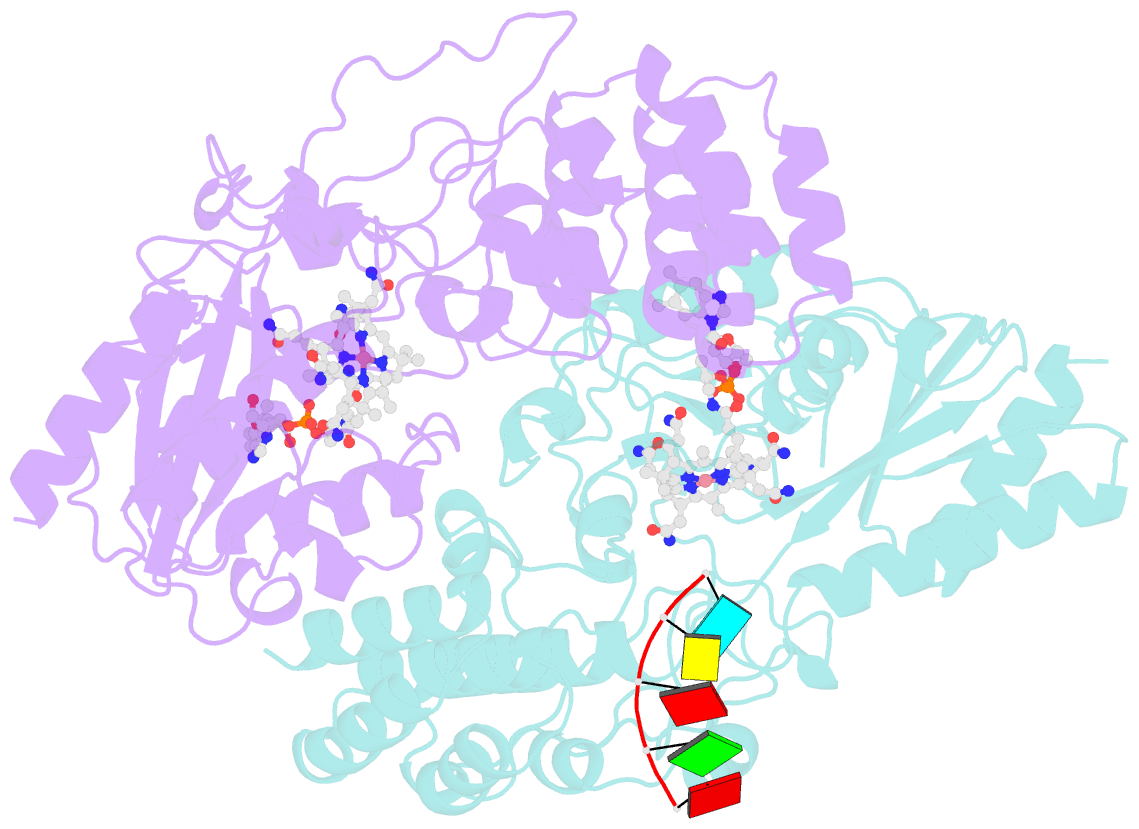
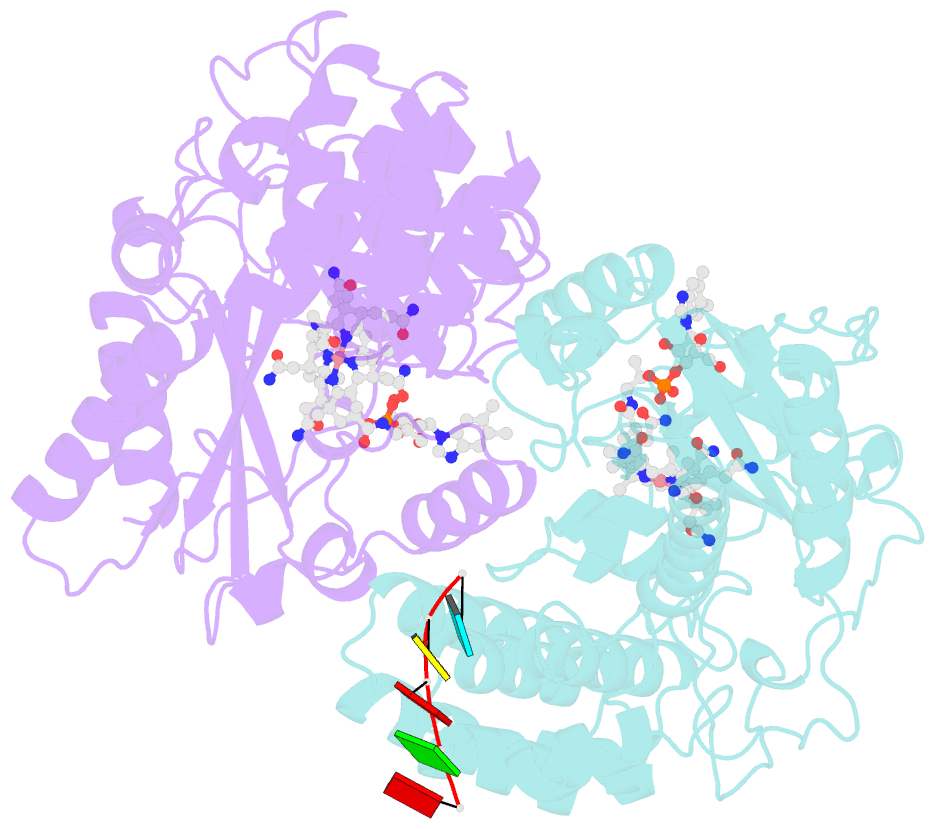
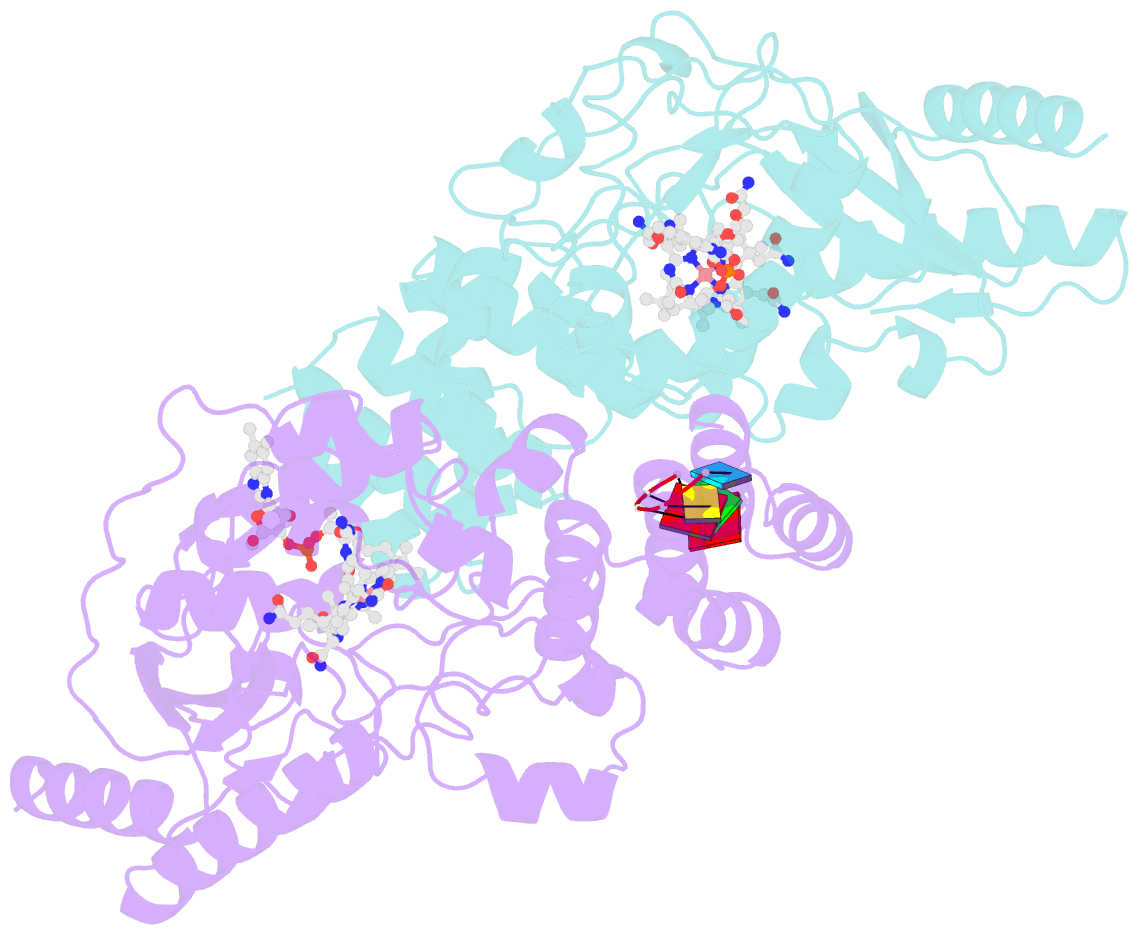
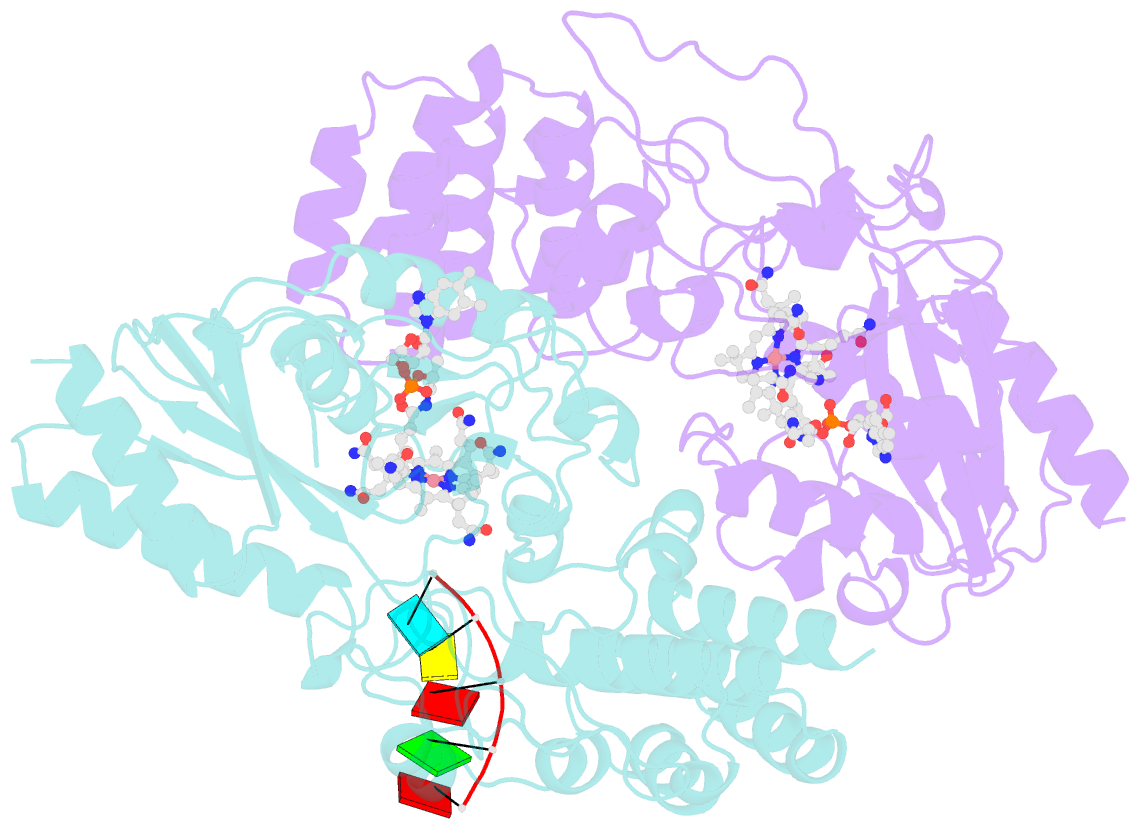
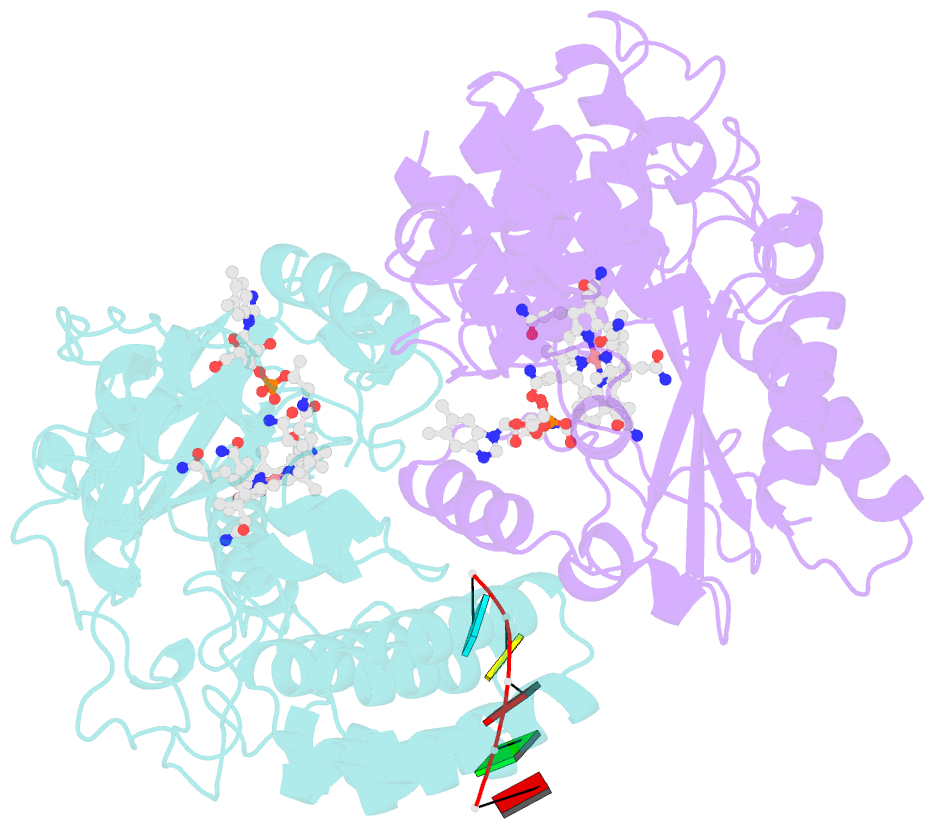
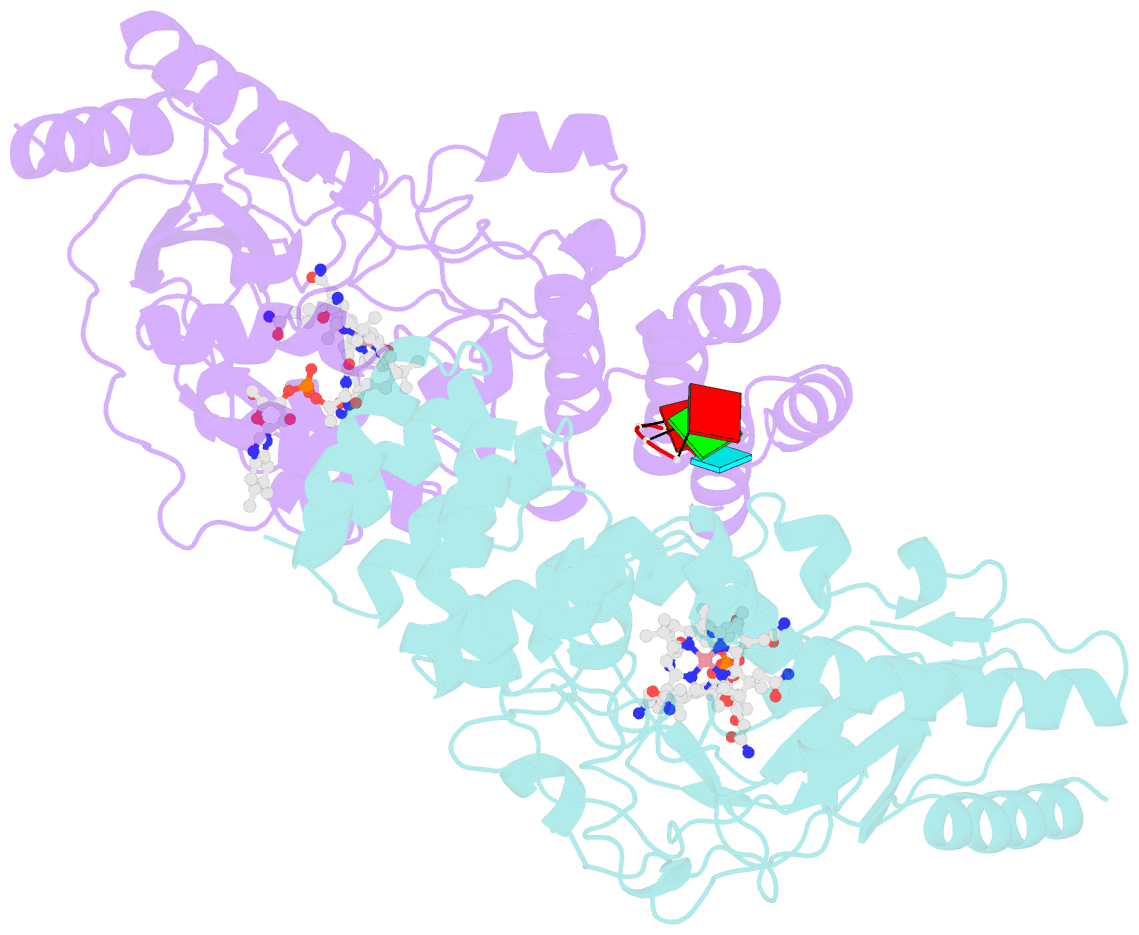
- Structure of epoxyqueuosine reductase from bacillus subtilis with the asp134 catalytic loop swung out of the active site.
- Reference
- Dowling DP, Miles ZD, Kohrer C, Maiocco SJ, Elliott SJ, Bandarian V, Drennan CL (2016): "Molecular basis of cobalamin-dependent RNA modification." Nucleic Acids Res., 44, 9965-9976. doi: 10.1093/nar/gkw806.
- Abstract
- Queuosine (Q) was discovered in the wobble position of a transfer RNA (tRNA) 47 years ago, yet the final biosynthetic enzyme responsible for Q-maturation, epoxyqueuosine (oQ) reductase (QueG), was only recently identified. QueG is a cobalamin (Cbl)-dependent, [4Fe-4S] cluster-containing protein that produces the hypermodified nucleoside Q in situ on four tRNAs. To understand how QueG is able to perform epoxide reduction, an unprecedented reaction for a Cbl-dependent enzyme, we have determined a series of high resolution structures of QueG from Bacillus subtilis Our structure of QueG bound to a tRNATyr anticodon stem loop shows how this enzyme uses a HEAT-like domain to recognize the appropriate anticodons and position the hypermodified nucleoside into the enzyme active site. We find Q bound directly above the Cbl, consistent with a reaction mechanism that involves the formation of a covalent Cbl-tRNA intermediate. Using protein film electrochemistry, we show that two [4Fe-4S] clusters adjacent to the Cbl have redox potentials in the range expected for Cbl reduction, suggesting how Cbl can be activated for nucleophilic attack on oQ. Together, these structural and electrochemical data inform our understanding of Cbl dependent nucleic acid modification.