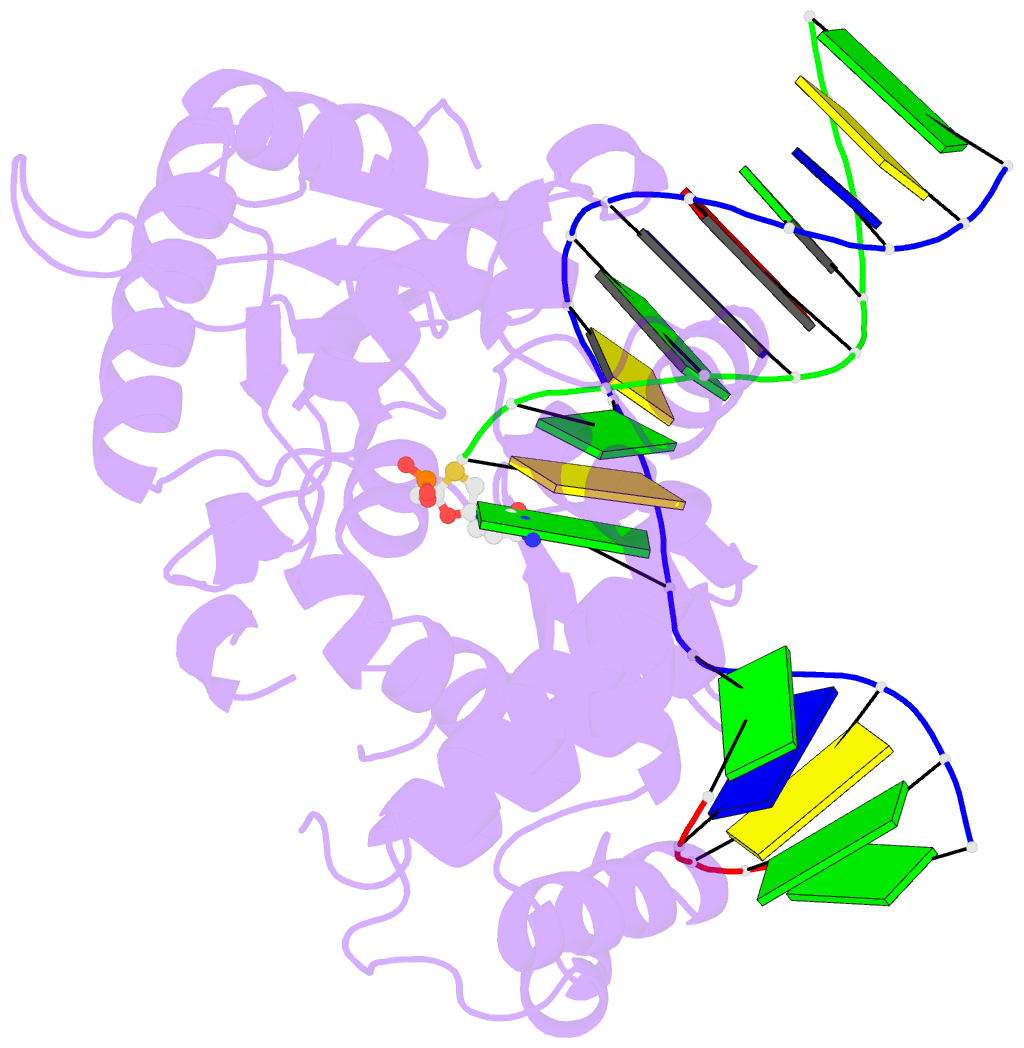
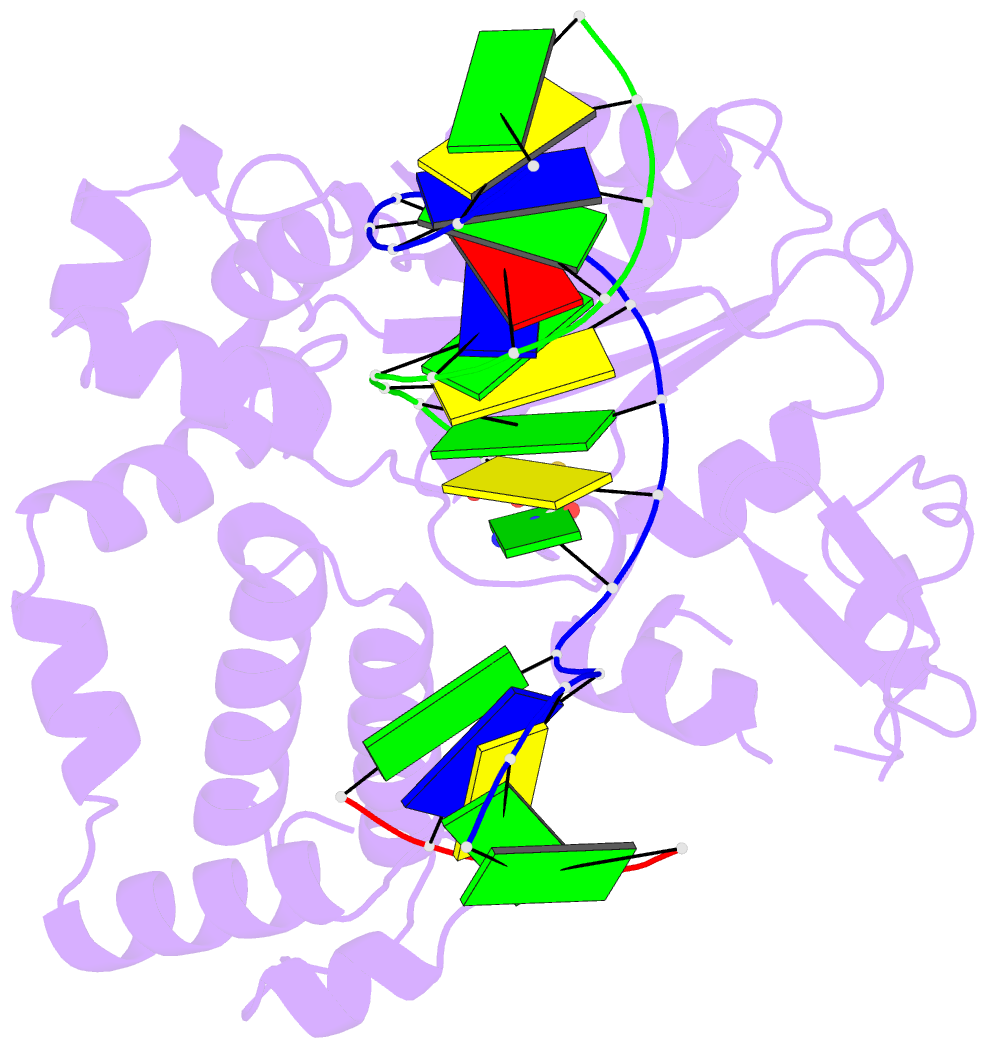
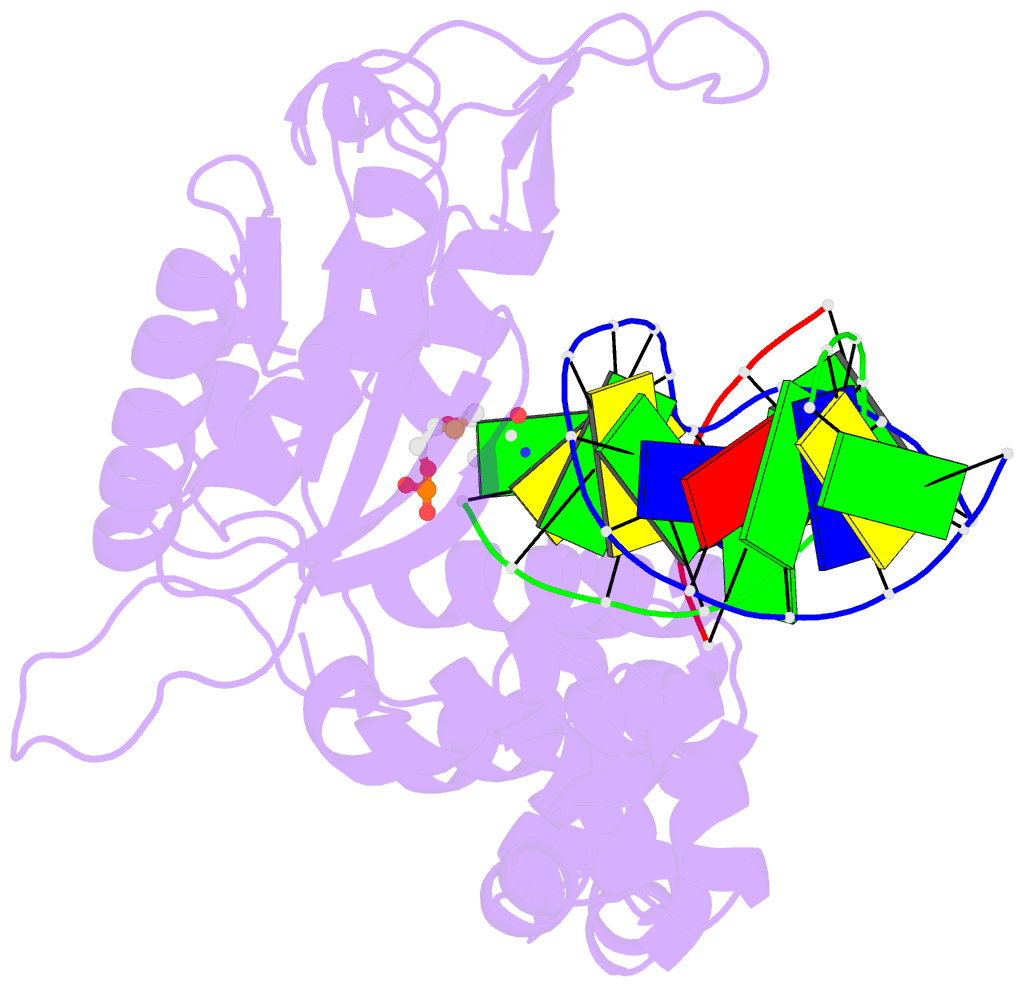
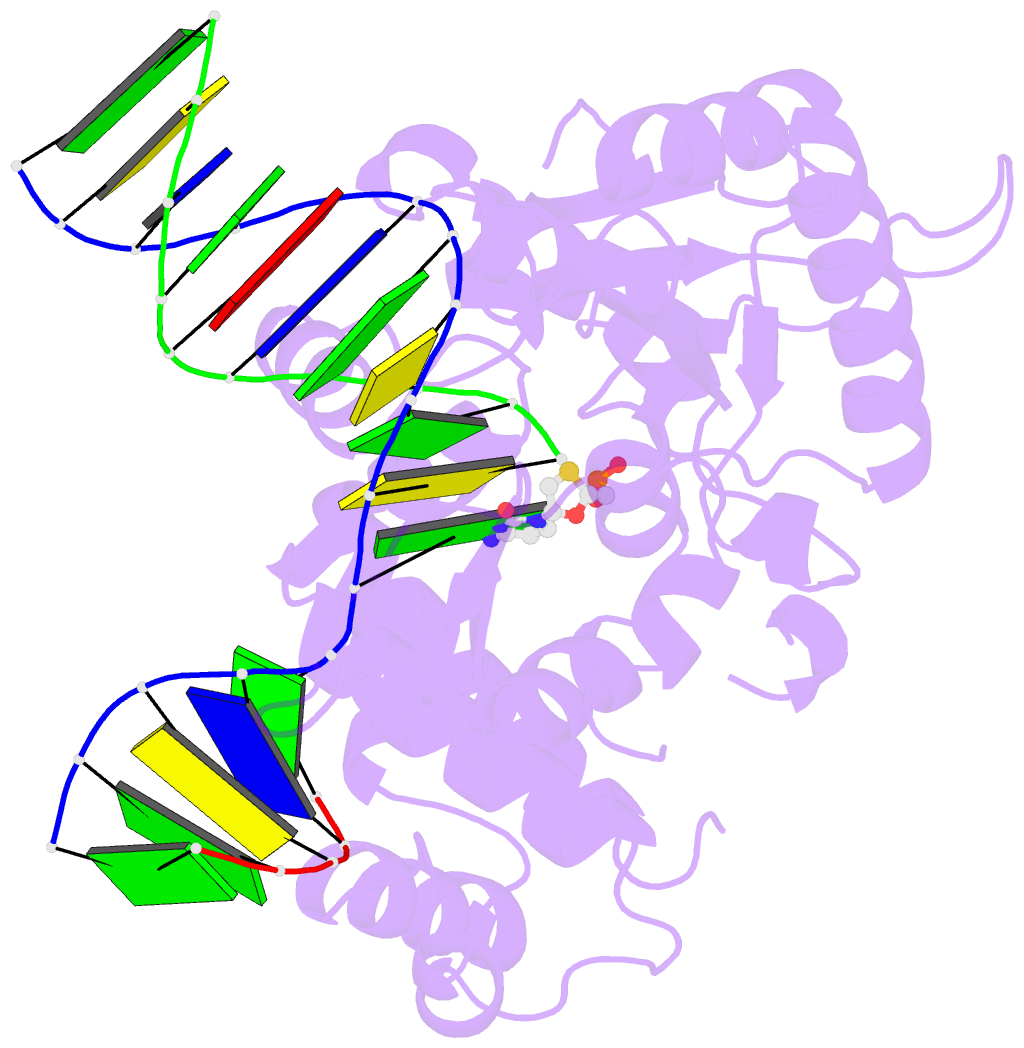
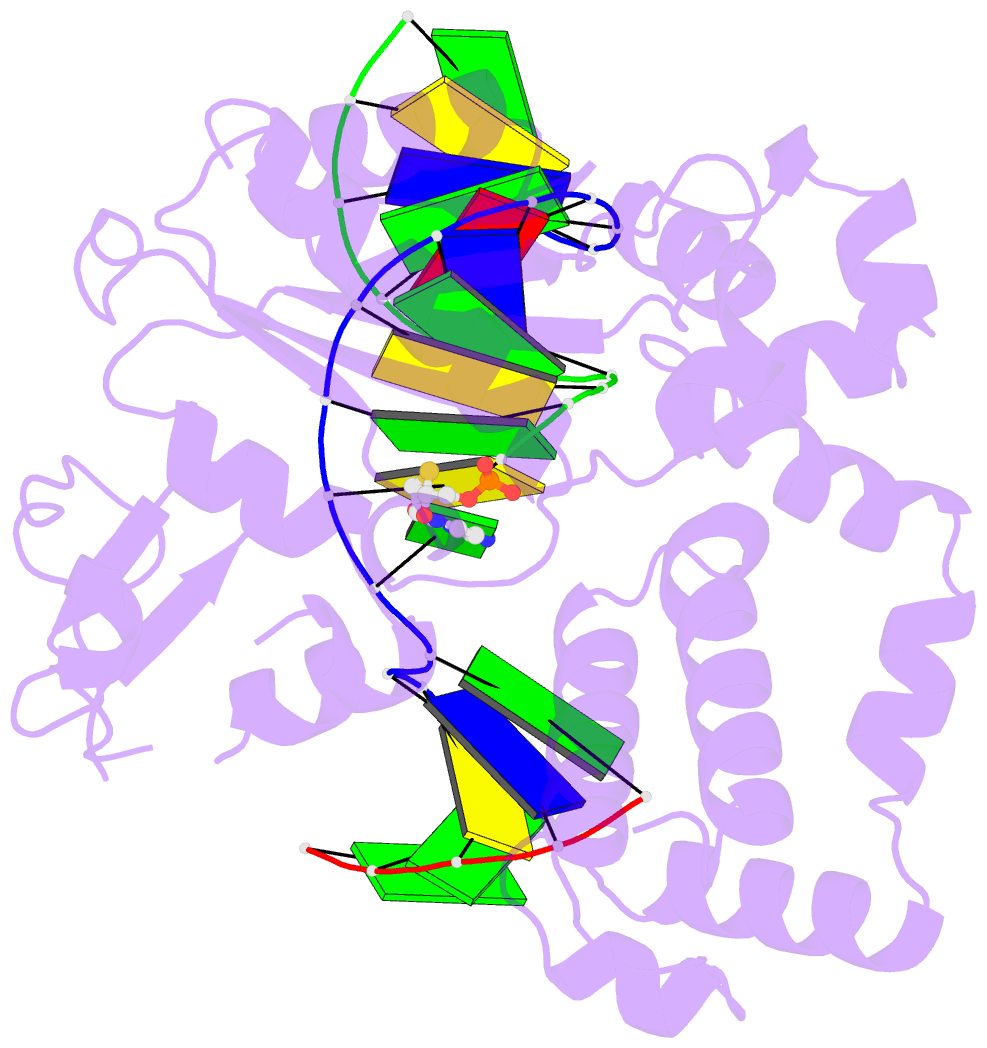
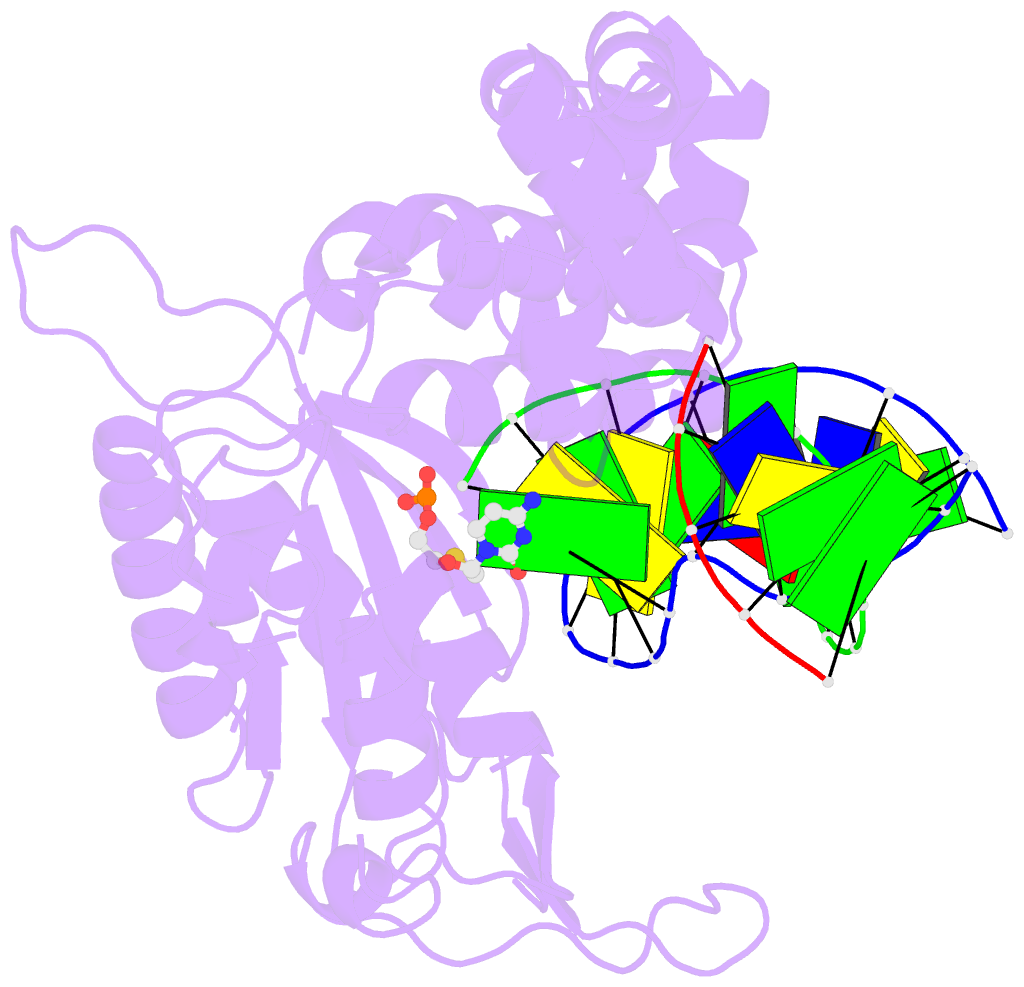
Summary information and primary citation
- PDB-id
- 5tba; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.49 Å)
- Summary
- Postcatalytic ternary complex of human DNA polymerase beta with gapped DNA substrate, incorporated (-)3tc and ppi.
- Reference
- Reed AJ, Vyas R, Raper AT, Suo Z (2017): "Structural Insights into the Post-Chemistry Steps of Nucleotide Incorporation Catalyzed by a DNA Polymerase." J. Am. Chem. Soc., 139, 465-471. doi: 10.1021/jacs.6b11258.
- Abstract
- DNA polymerases are essential enzymes that faithfully and efficiently replicate genomic information.1-3 The mechanism of nucleotide incorporation by DNA polymerases has been extensively studied structurally and kinetically, but several key steps following phosphodiester bond formation remain structurally uncharacterized due to utilization of natural nucleotides. It is thought that the release of pyrophosphate (PPi) triggers reverse conformational changes in a polymerase in order to complete a full catalytic cycle as well as prepare for DNA translocation and subsequent incorporation events. Here, by using the triphosphates of chain-terminating antiviral drugs lamivudine ((-)3TC-TP) and emtricitabine ((-)FTC-TP), we structurally reveal the correct sequence of post-chemistry steps during nucleotide incorporation by human DNA polymerase β (hPolβ) and provide a structural basis for PPi release. These post-catalytic structures reveal hPolβ in an open conformation with PPi bound in the active site, thereby strongly suggesting that the reverse conformational changes occur prior to PPi release. The results also help to refine the role of the newly discovered third divalent metal ion for DNA polymerase-catalyzed nucleotide incorporation. Furthermore, a post-chemistry structure of hPolβ in the open conformation, following incorporation of (-)3TC-MP, with a second (-)3TC-TP molecule bound to the active site in the absence of PPi, suggests that nucleotide binding stimulates PPi dissociation and occurs before polymerase translocation. Our structural characterization defines the order of the elusive post-chemistry steps in the canonical mechanism of a DNA polymerase.