Summary information and primary citation
- PDB-id
- 5tgx; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- X-ray (2.3 Å)
- Summary
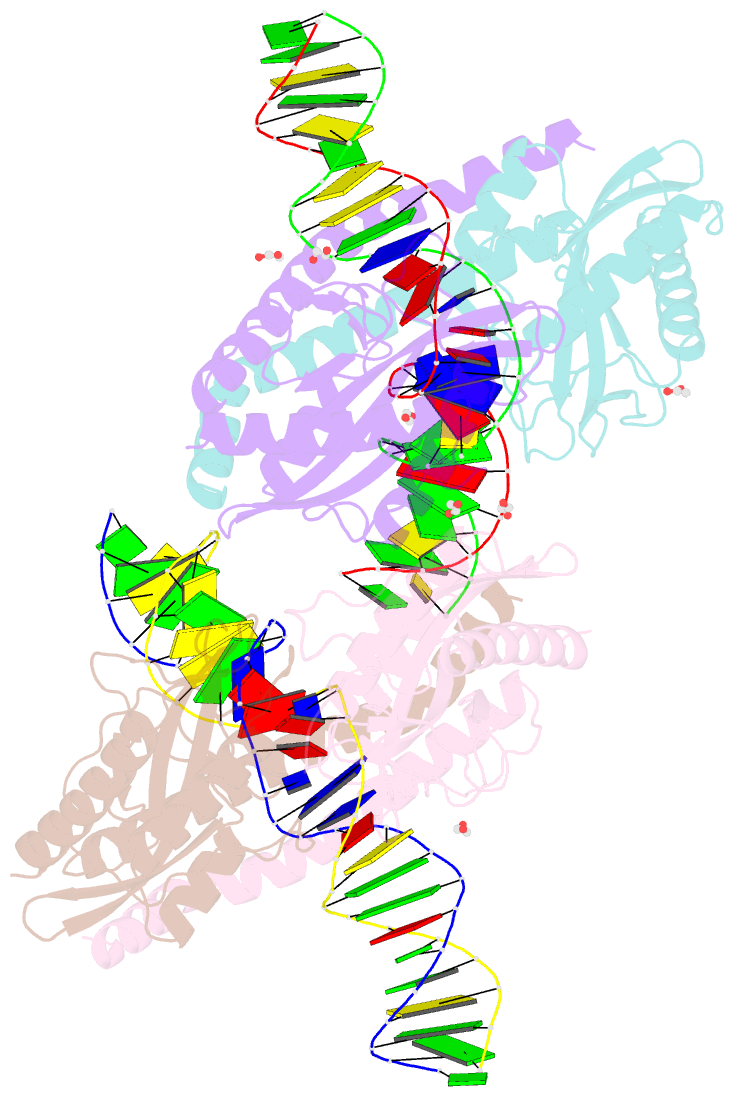
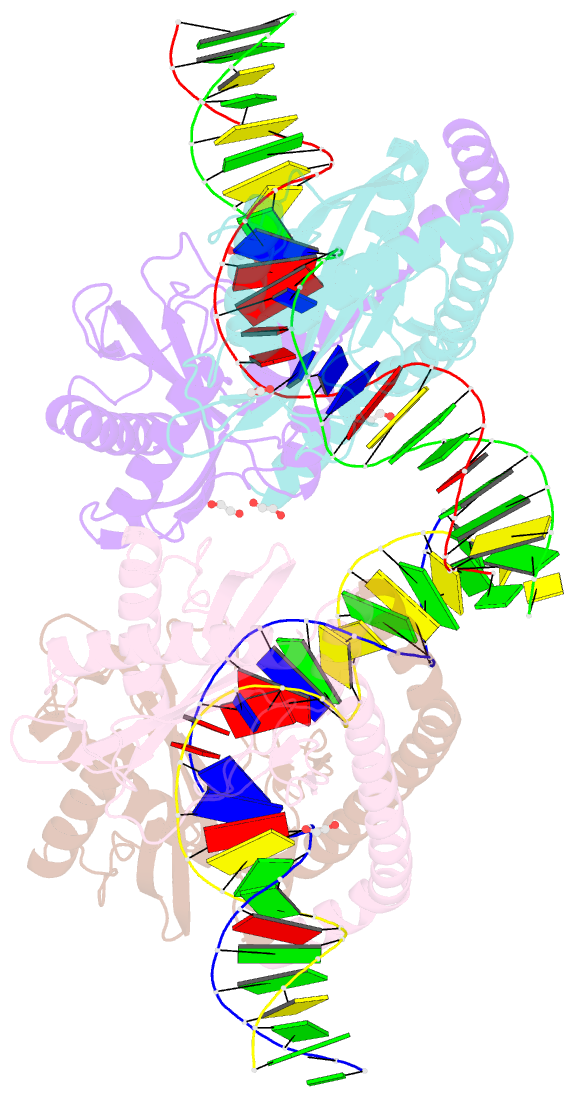
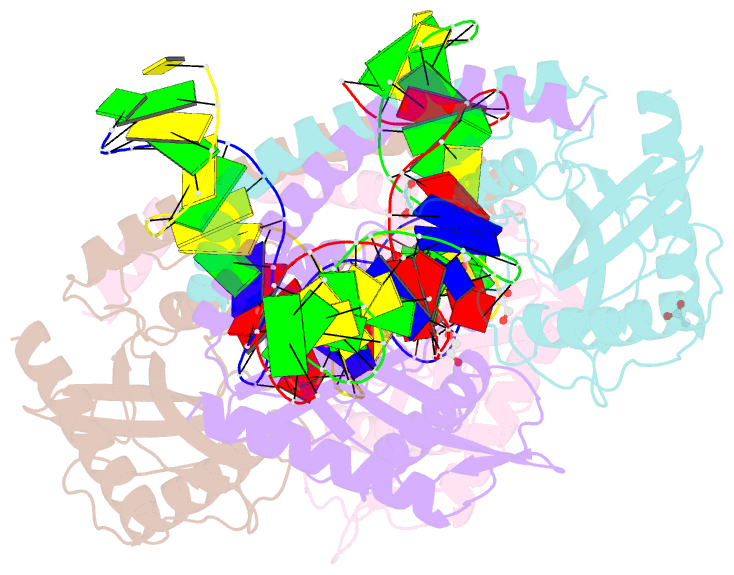
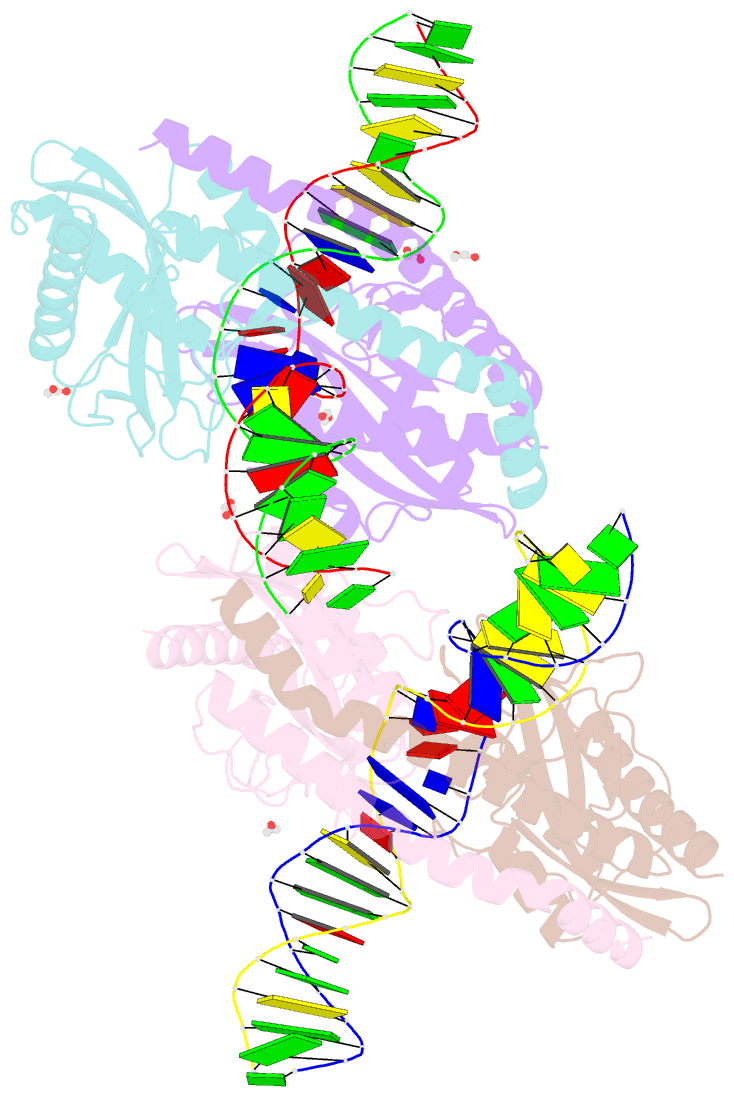
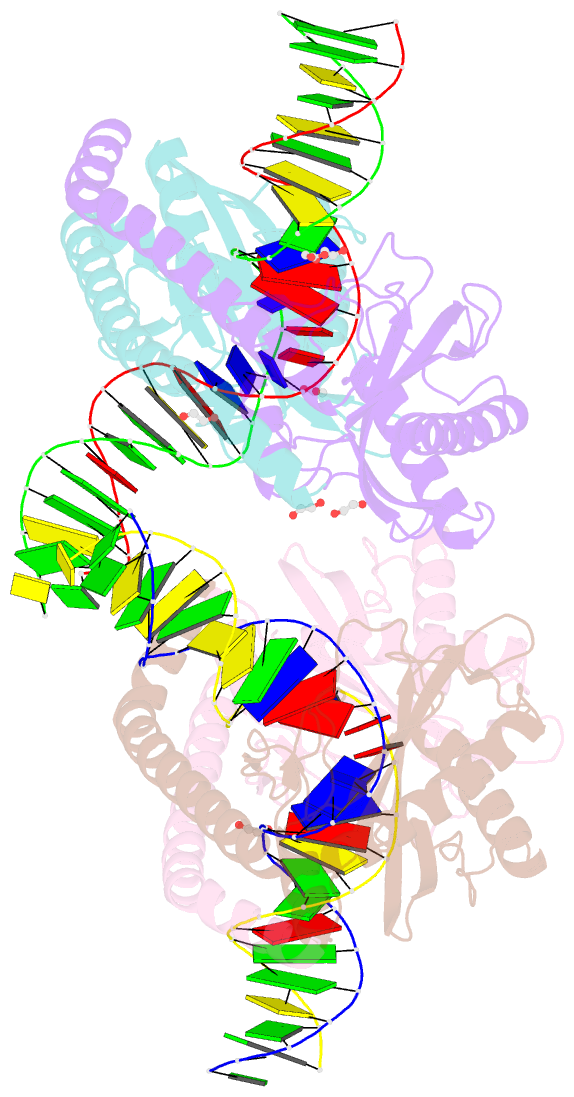
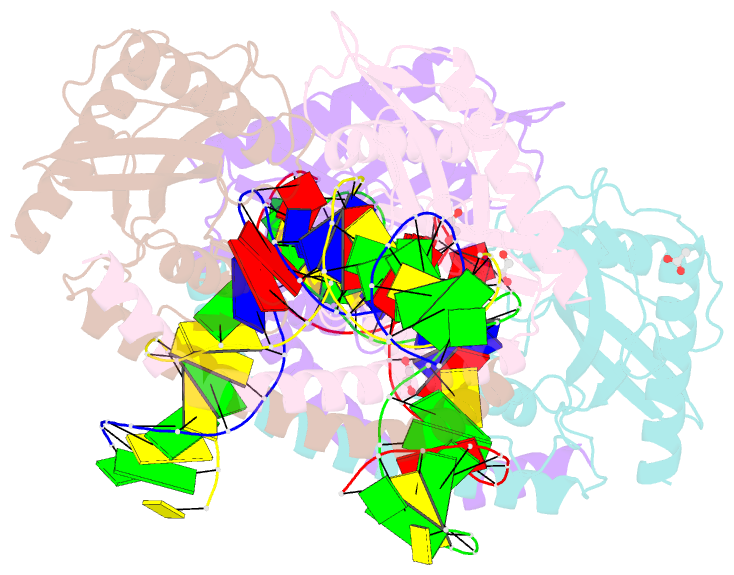
- Restriction-modification system-type ii r-swai complexed with partially cleaved DNA
- Reference
- Shen BW, Heiter DF, Lunnen KD, Wilson GG, Stoddard BL (2017): "DNA recognition by the SwaI restriction endonuclease involves unusual distortion of an 8 base pair A:T-rich target." Nucleic Acids Res., 45, 1516-1528. doi: 10.1093/nar/gkw1200.
- Abstract
- R.SwaI, a Type IIP restriction endonuclease, recognizes a palindromic eight base pair (bp) symmetric sequence, 5΄-ATTTAAAT-3΄, and cleaves that target at its center to generate blunt-ended DNA fragments. Here, we report three crystal structures of SwaI: unbound enzyme, a DNA-bound complex with calcium ions; and a DNA-bound, fully cleaved complex with magnesium ions. We compare these structures to two structurally similar ‘PD-D/ExK’ restriction endonucleases (EcoRV and HincII) that also generate blunt-ended products, and to a structurally distinct enzyme (the HNH endonuclease PacI) that also recognizes an 8-bp target site consisting solely of A:T base pairs. Binding by SwaI induces an extreme bend in the target sequence accompanied by un-pairing and re-ordering of its central A:T base pairs. This result is reminiscent of a more dramatic target deformation previously described for PacI, implying that long A:T-rich target sites might display structural or dynamic behaviors that play a significant role in endonuclease recognition and cleavage.