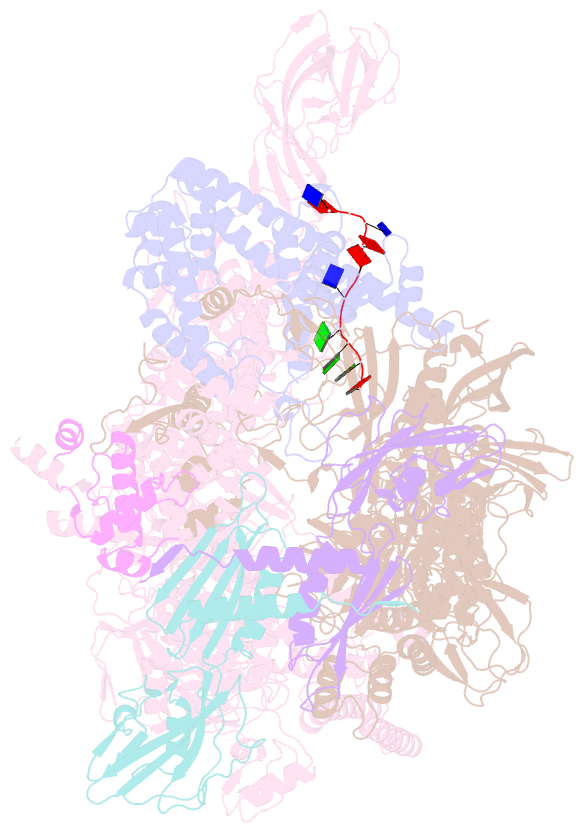
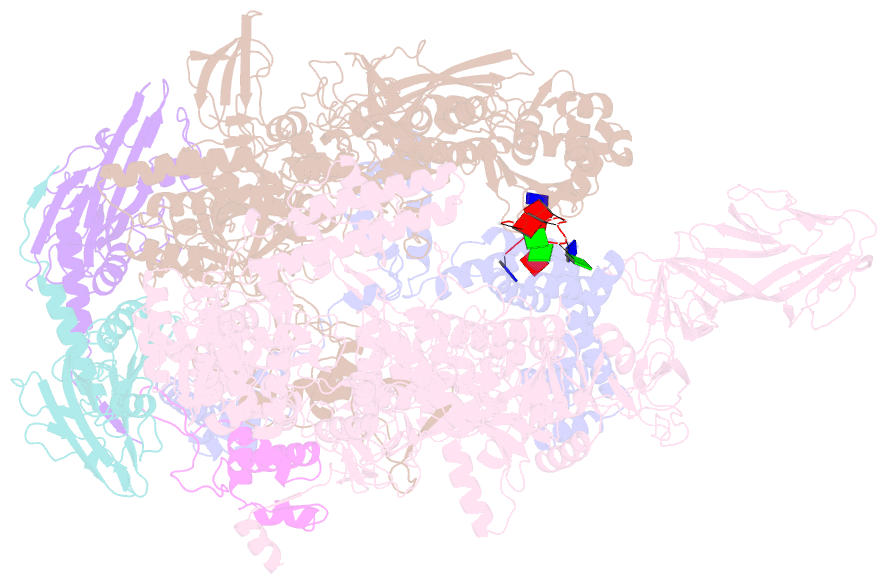
Summary information and primary citation
- PDB-id
- 5tjg; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.6 Å)
- Summary
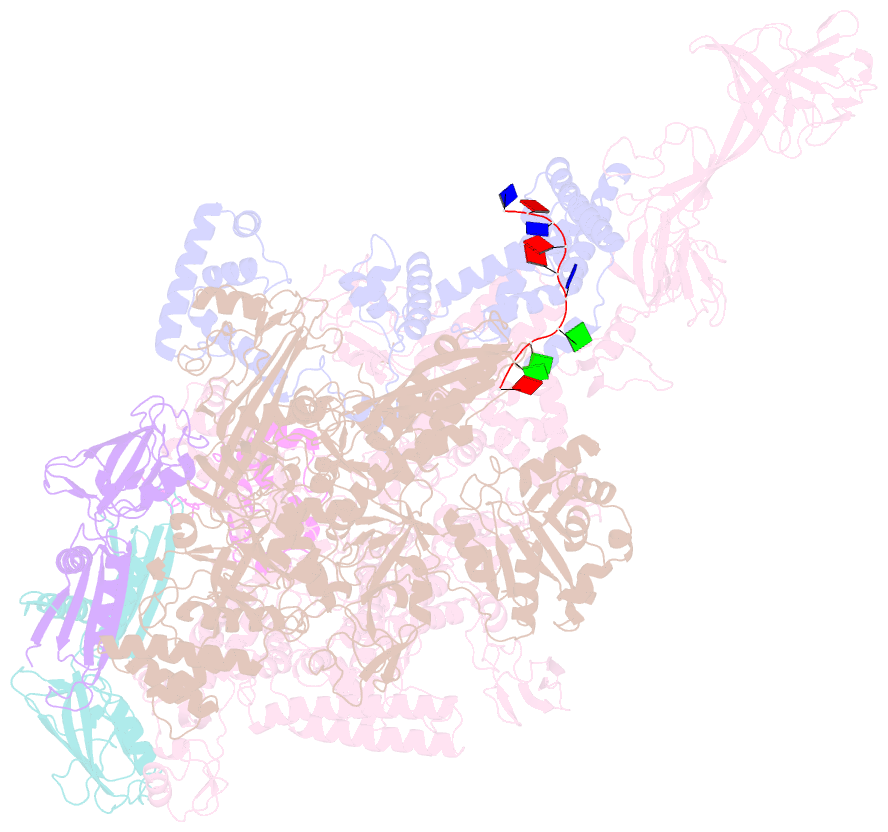
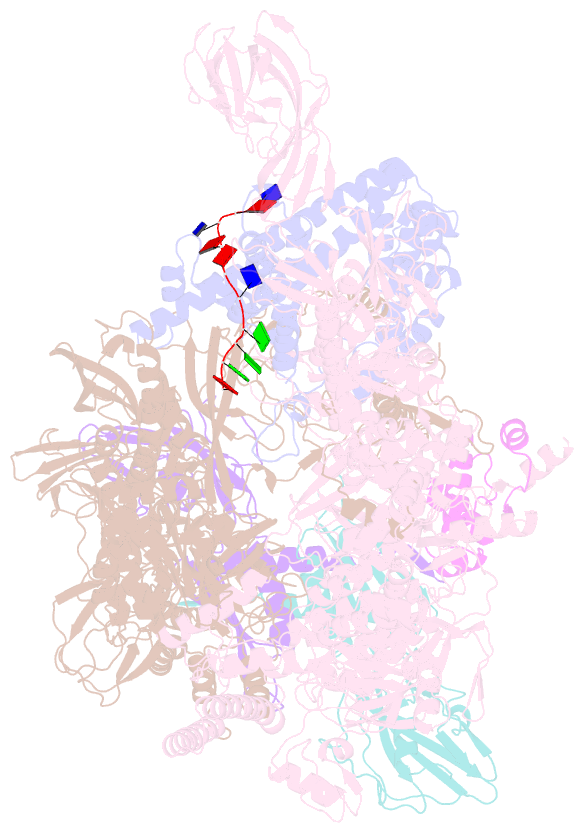
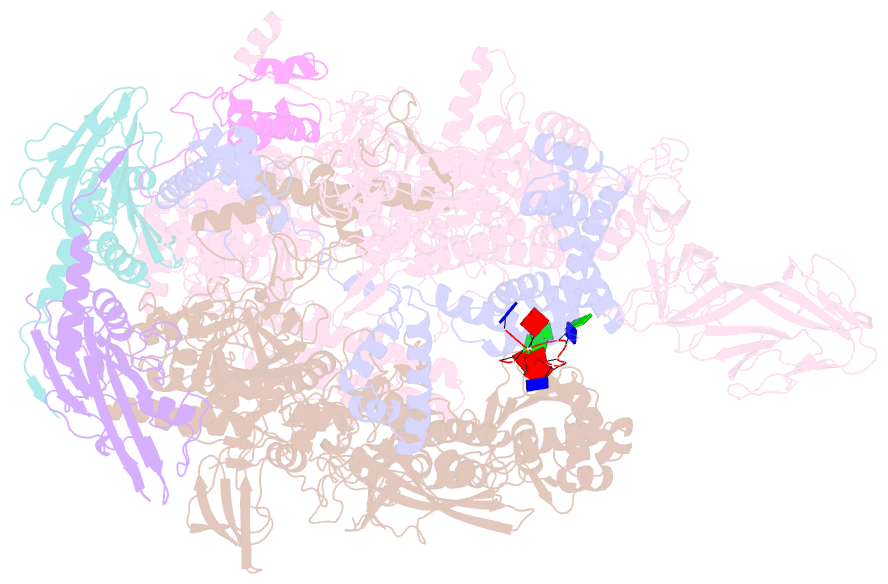
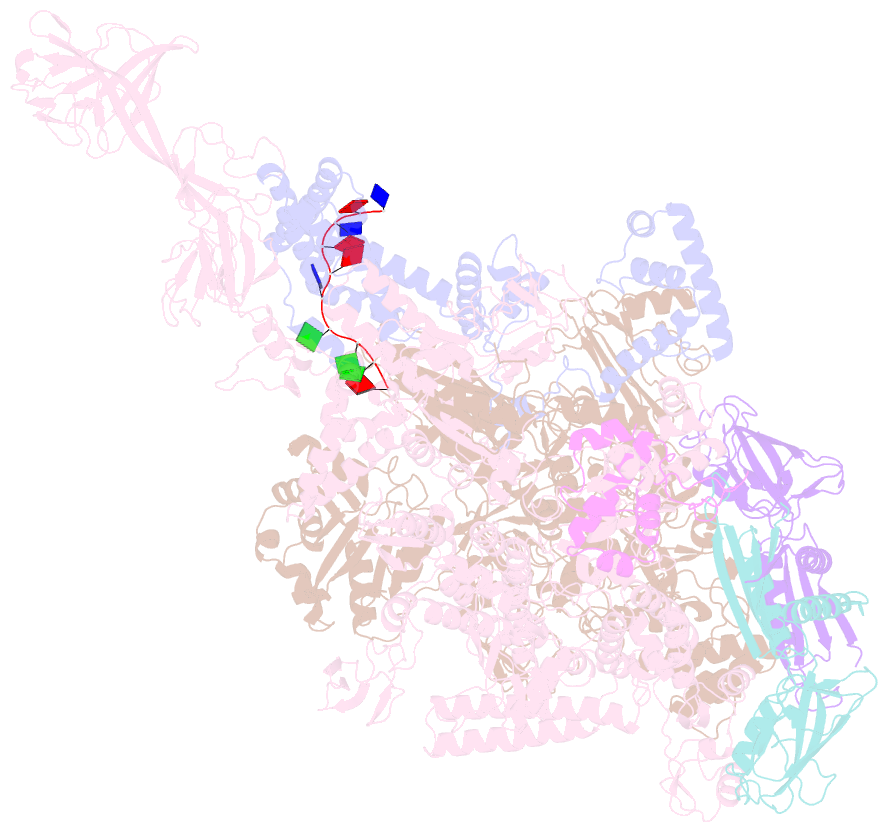
- Thermus aquaticus delta1.1-sigmaa holoenzyme-downstream-fork promoter complex with an open clamp
- Reference
- Feklistov A, Bae B, Hauver J, Lass-Napiorkowska A, Kalesse M, Glaus F, Altmann KH, Heyduk T, Landick R, Darst SA (2017): "RNA polymerase motions during promoter melting." Science, 356, 863-866. doi: 10.1126/science.aam7858.
- Abstract
- All cellular RNA polymerases (RNAPs), from those of bacteria to those of man, possess a clamp that can open and close, and it has been assumed that the open RNAP separates promoter DNA strands and then closes to establish a tight grip on the DNA template. Here, we resolve successive motions of the initiating bacterial RNAP by studying real-time signatures of fluorescent reporters placed on RNAP and DNA in the presence of ligands locking the clamp in distinct conformations. We report evidence for an unexpected and obligatory step early in the initiation involving a transient clamp closure as a prerequisite for DNA melting. We also present a 2.6-angstrom crystal structure of a late-initiation intermediate harboring a rotationally unconstrained downstream DNA duplex within the open RNAP active site cleft. Our findings explain how RNAP thermal motions control the promoter search and drive DNA melting in the absence of external energy sources.