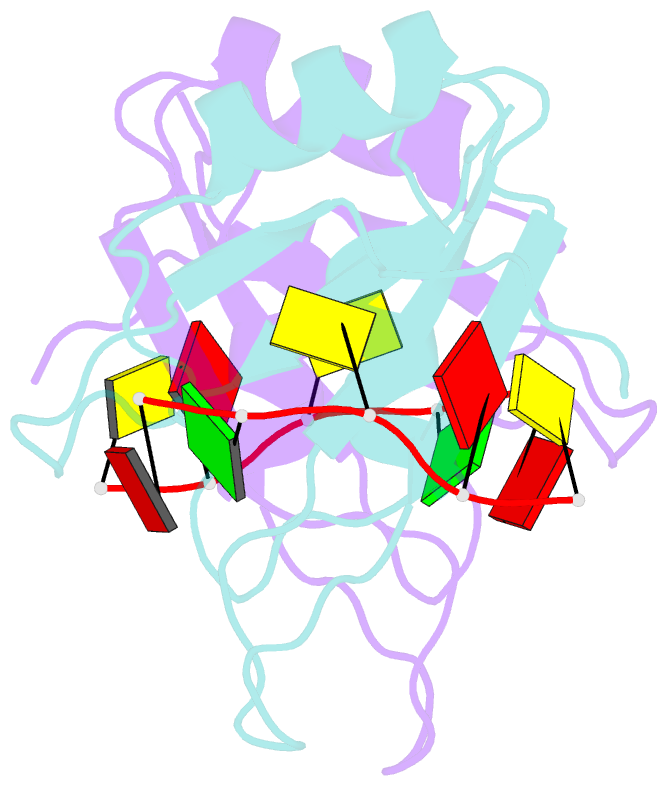
Summary information and primary citation
- PDB-id
- 5tkz; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- splicing
- Method
- X-ray (1.529 Å)
- Summary
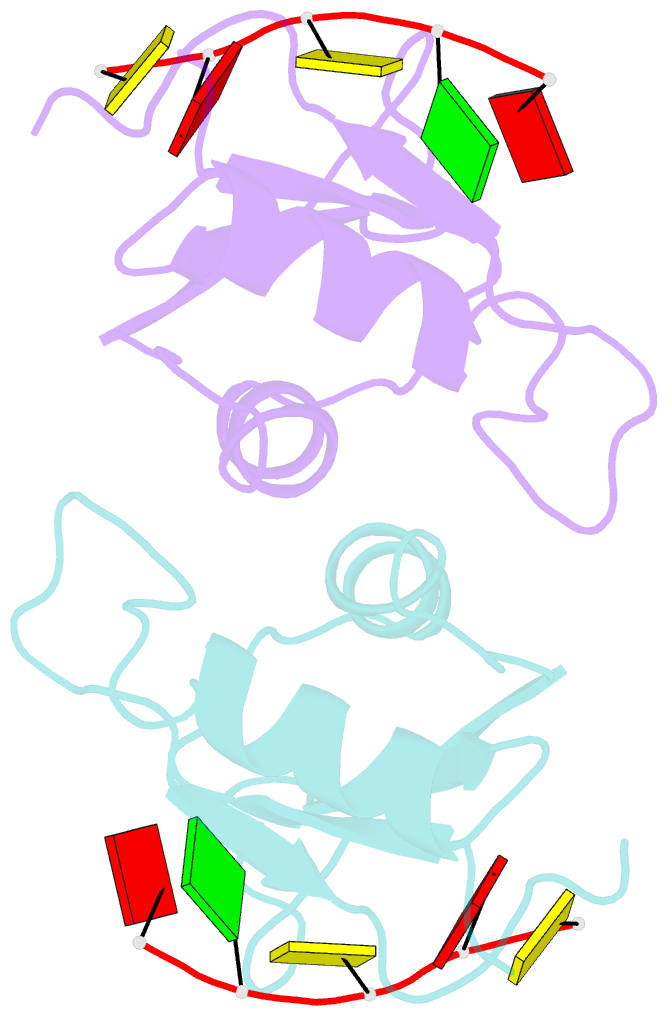
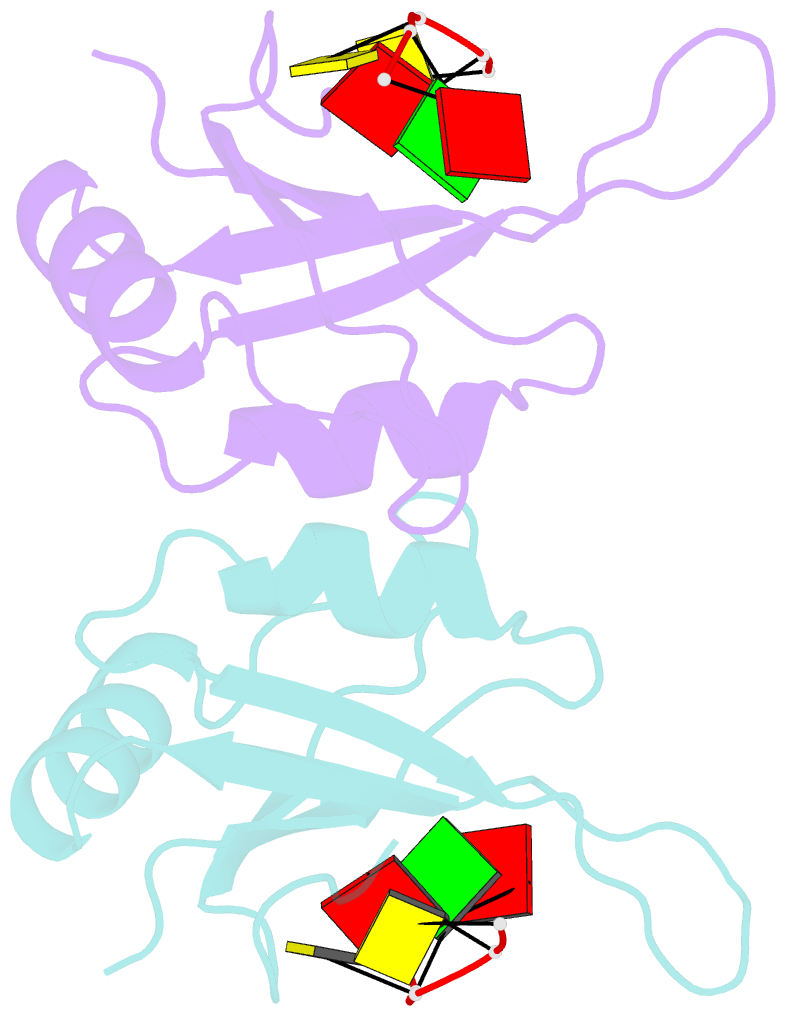
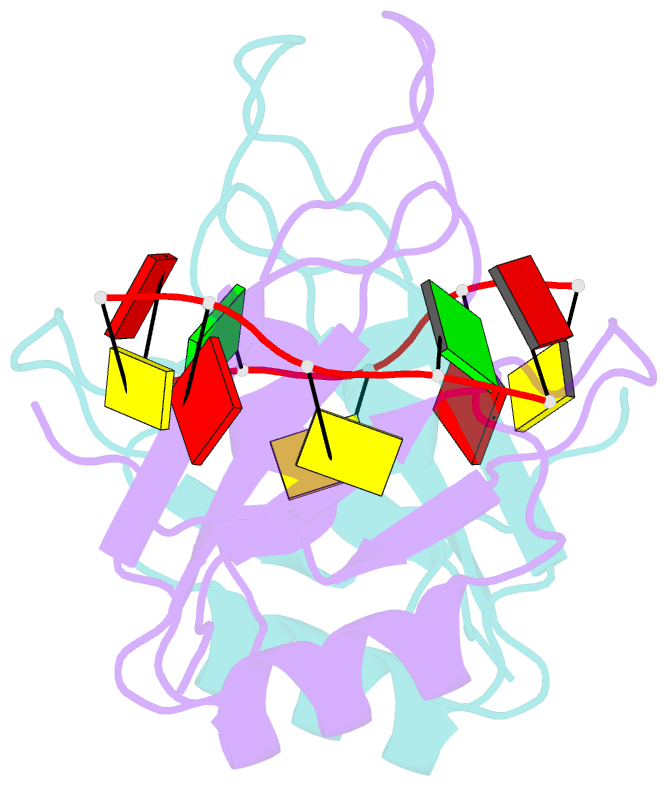
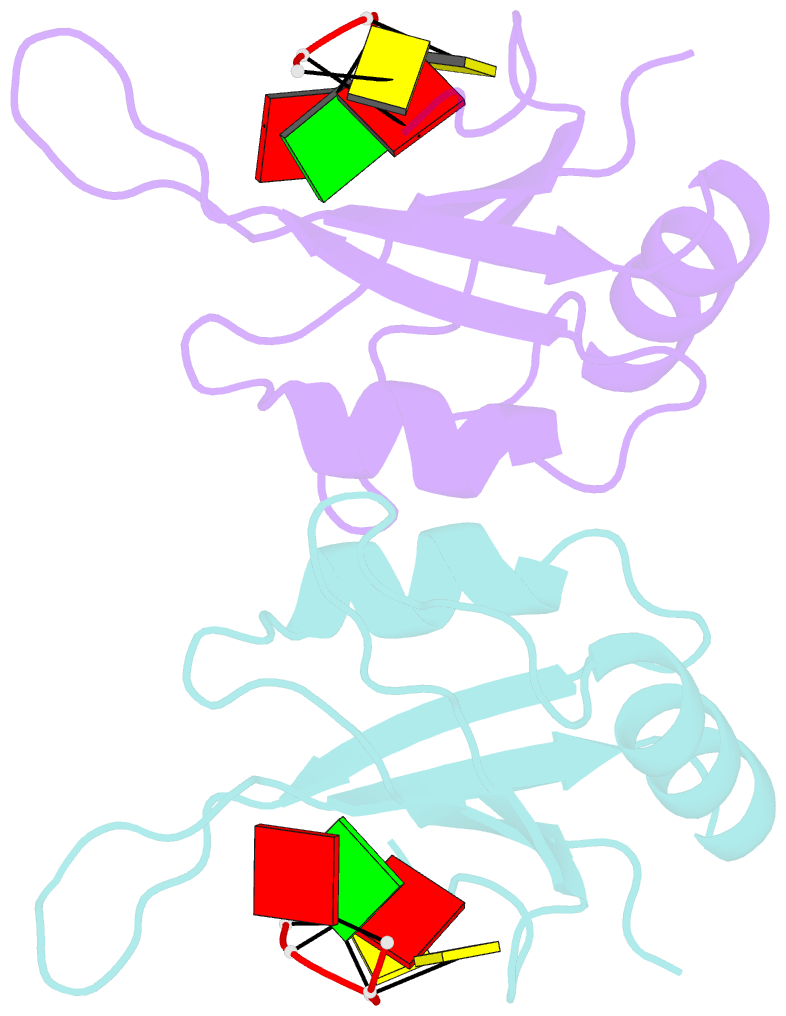
- Mec-8 n-terminal rrm bound to tandem gcac ligand
- Reference
- Soufari H, Mackereth CD (2017): "Conserved binding of GCAC motifs by MEC-8, couch potato, and the RBPMS protein family." RNA, 23, 308-316. doi: 10.1261/rna.059733.116.
- Abstract
- Precise regulation of mRNA processing, translation, localization, and stability relies on specific interactions with RNA-binding proteins whose biological function and target preference are dictated by their preferred RNA motifs. The RBPMS family of RNA-binding proteins is defined by a conserved RNA recognition motif (RRM) domain found in metazoan RBPMS/Hermes and RBPMS2, Drosophila couch potato, and MEC-8 from Caenorhabditis elegans In order to determine the parameters of RNA sequence recognition by the RBPMS family, we have first used the N-terminal domain from MEC-8 in binding assays and have demonstrated a preference for two GCAC motifs optimally separated by >6 nucleotides (nt). We have also determined the crystal structure of the dimeric N-terminal RRM domain from MEC-8 in the unbound form, and in complex with an oligonucleotide harboring two copies of the optimal GCAC motif. The atomic details reveal the molecular network that provides specificity to all four bases in the motif, including multiple hydrogen bonds to the initial guanine. Further studies with human RBPMS, as well as Drosophila couch potato, confirm a general preference for this double GCAC motif by other members of the protein family and the presence of this motif in known targets.