Summary information and primary citation
- PDB-id
- 5tzs; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- translation
- Method
- cryo-EM (5.1 Å)
- Summary
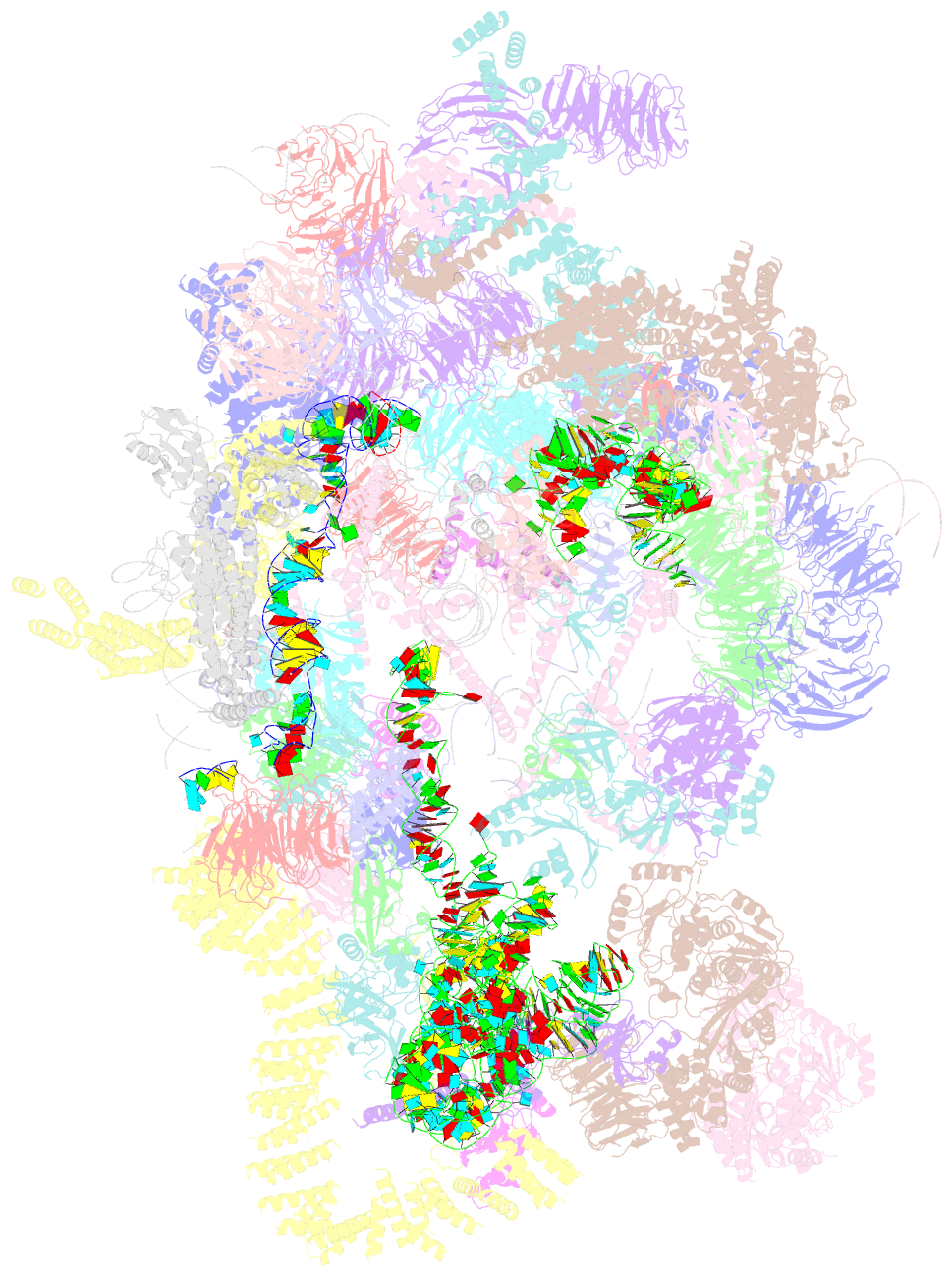
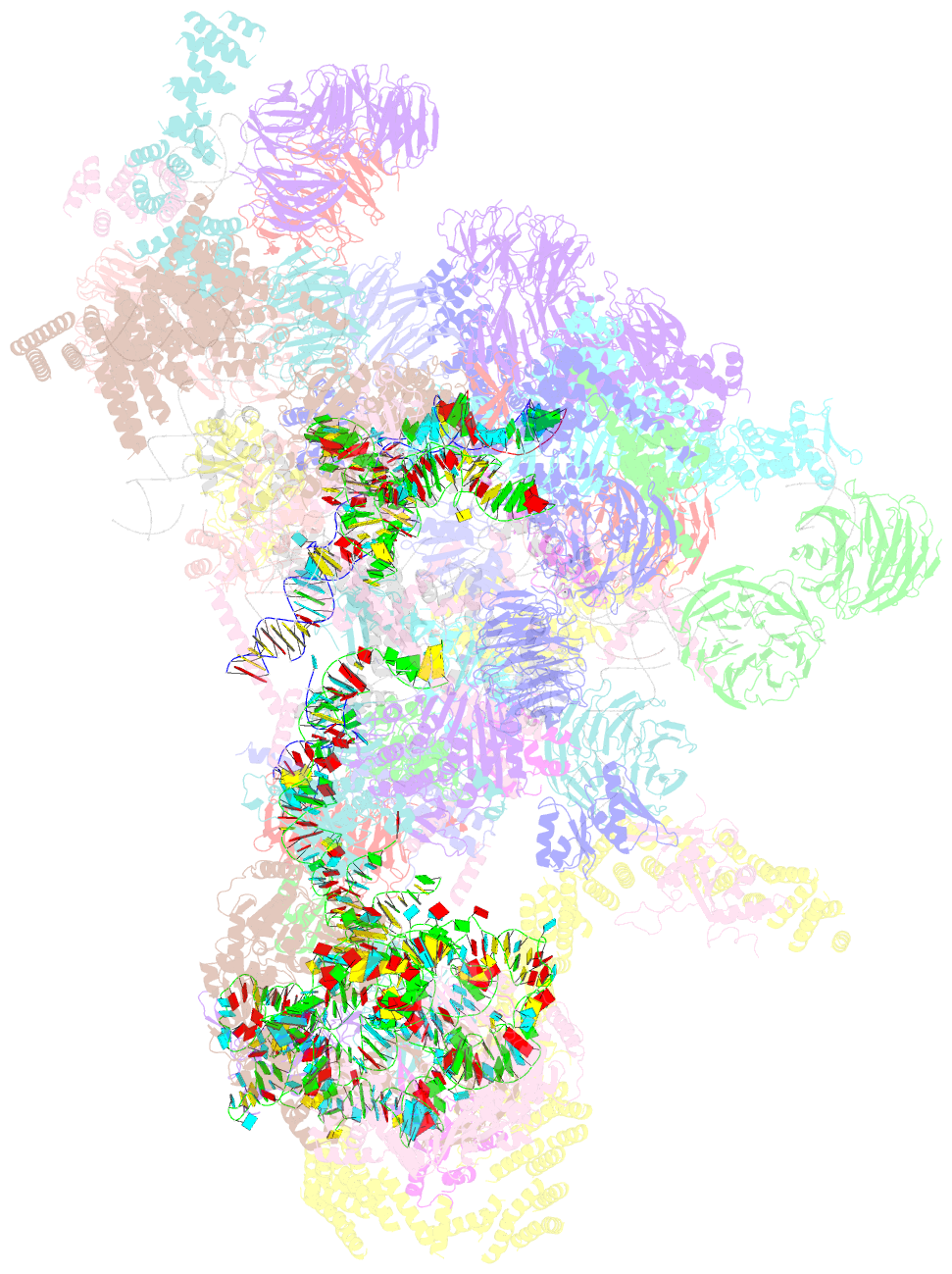
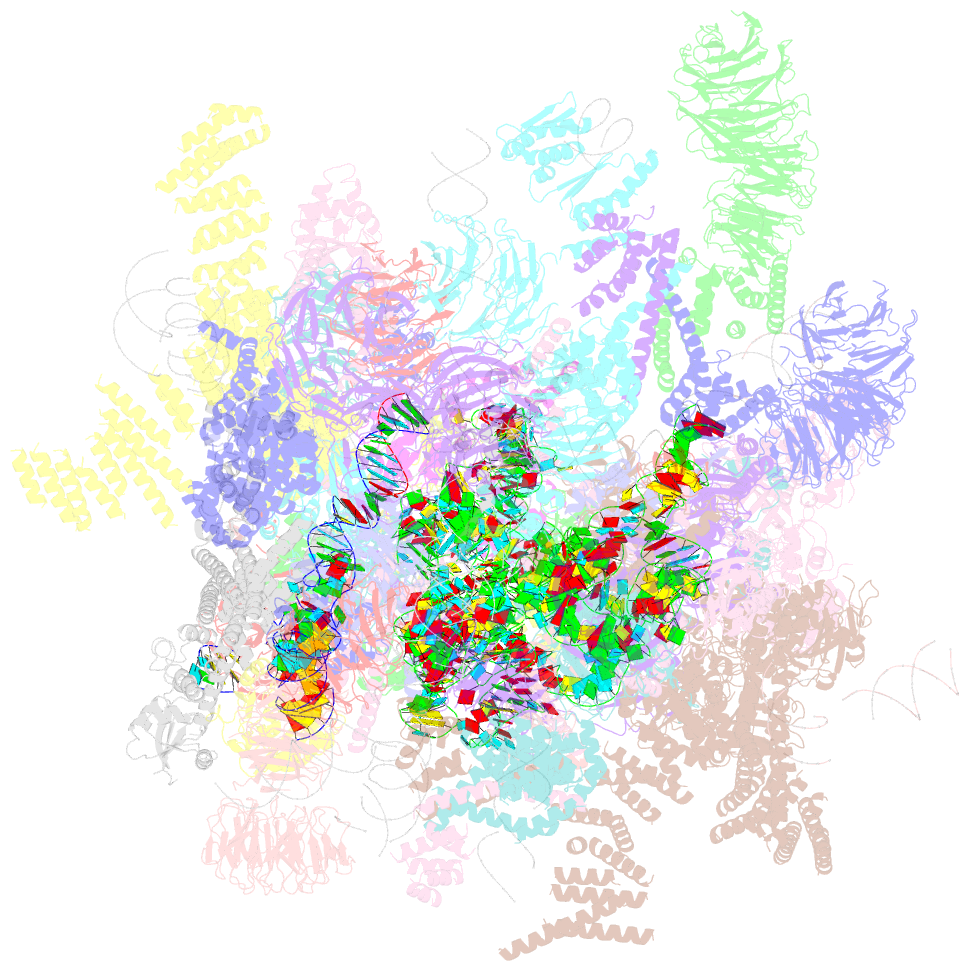
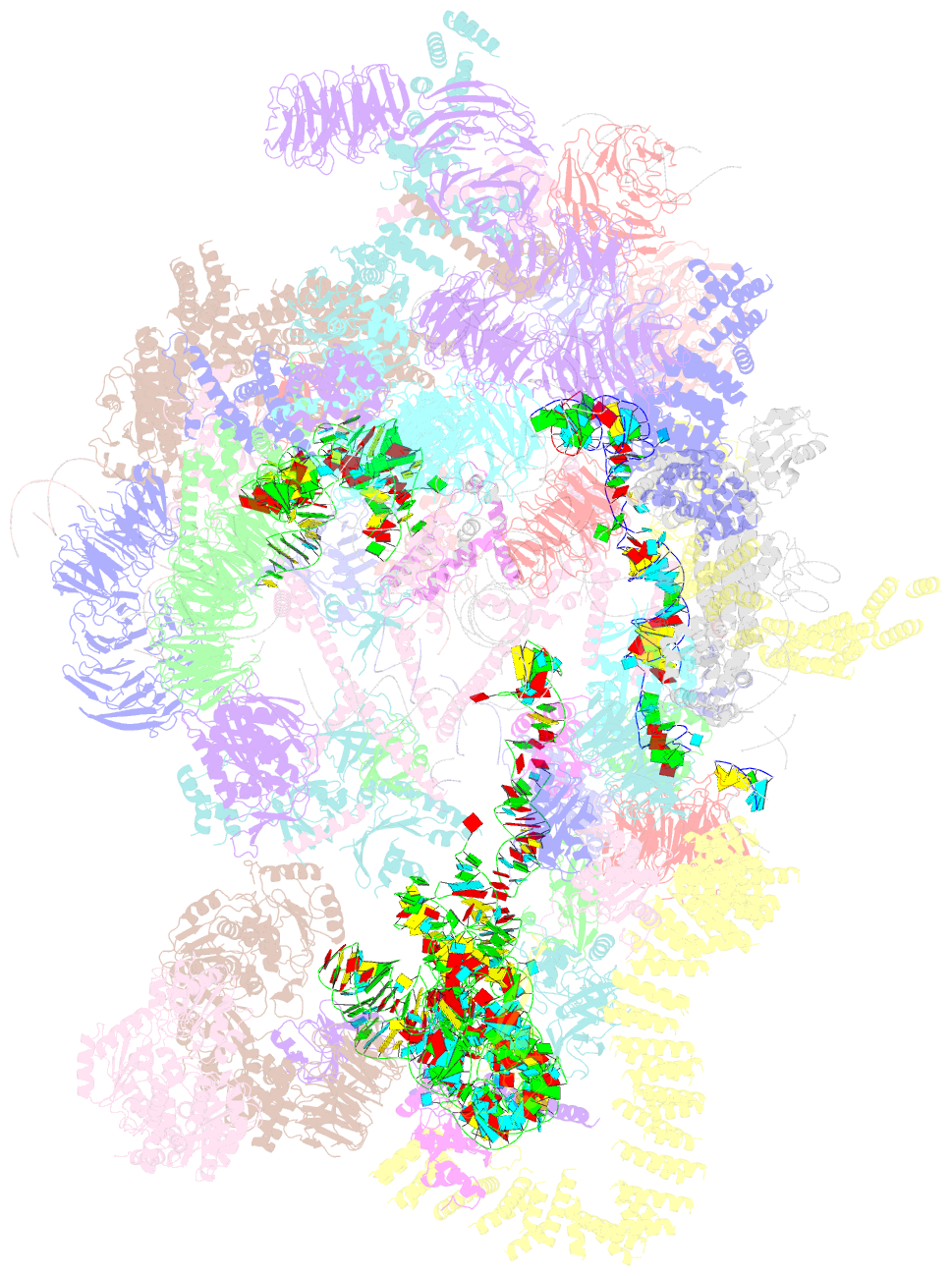
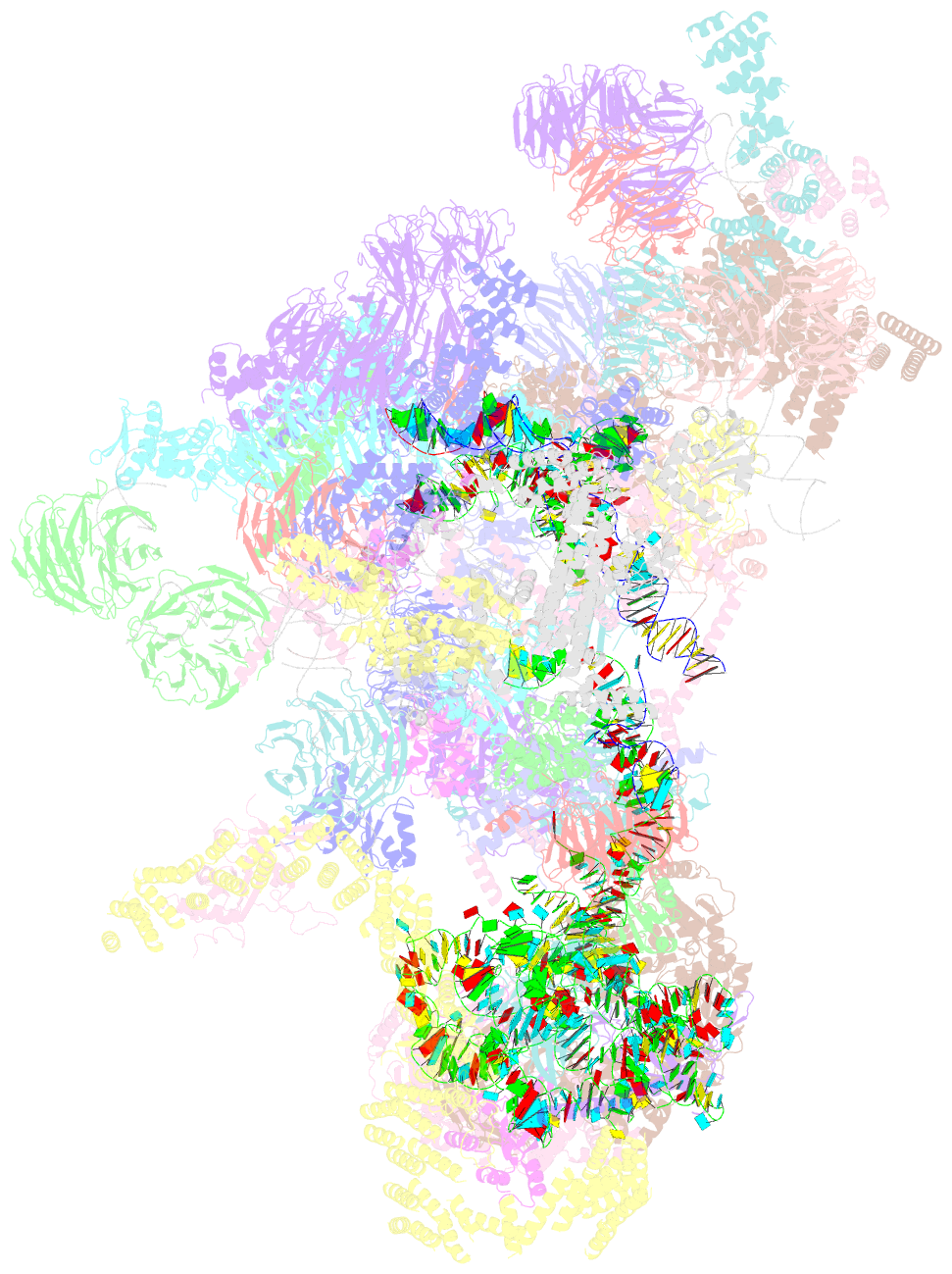
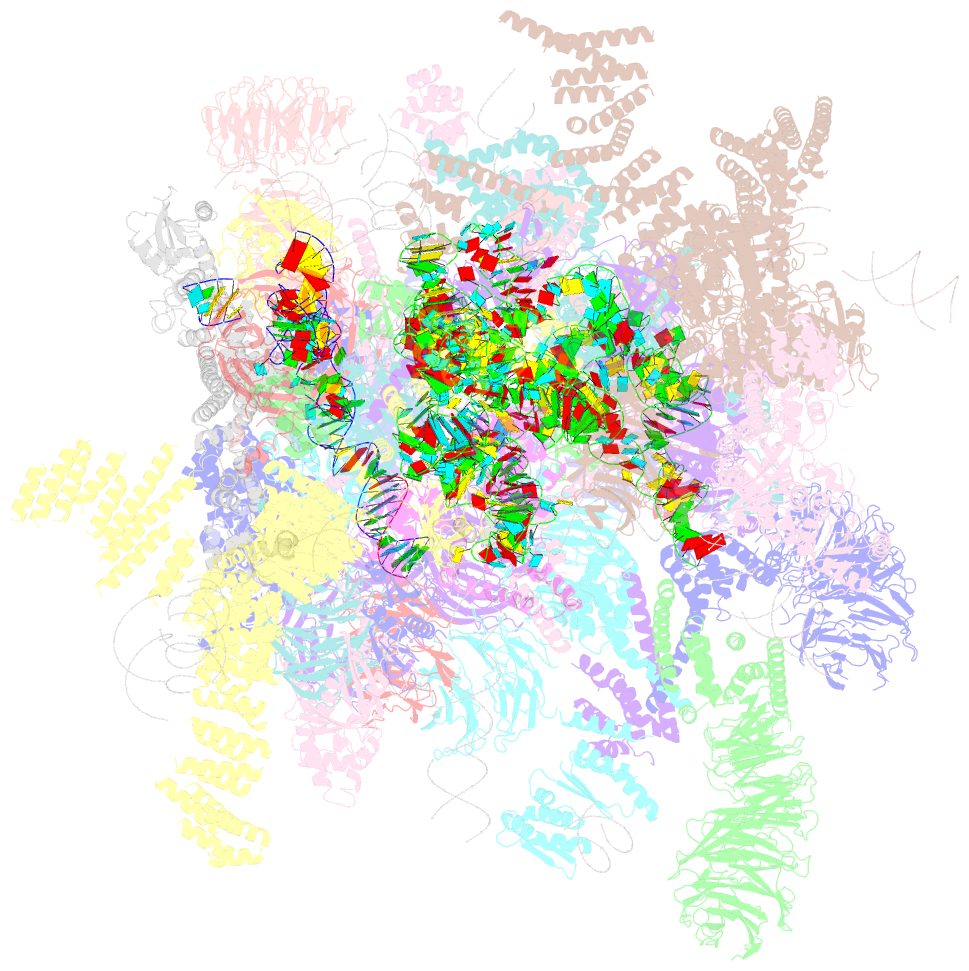
- Architecture of the yeast small subunit processome
- Reference
- Chaker-Margot M, Barandun J, Hunziker M, Klinge S (2017): "Architecture of the yeast small subunit processome." Science, 355. doi: 10.1126/science.aal1880.
- Abstract
- The small subunit (SSU) processome, a large ribonucleoprotein particle, organizes the assembly of the eukaryotic small ribosomal subunit by coordinating the folding, cleavage, and modification of nascent pre-ribosomal RNA (rRNA). Here, we present the cryo-electron microscopy structure of the yeast SSU processome at 5.1-angstrom resolution. The structure reveals how large ribosome biogenesis complexes assist the 5' external transcribed spacer and U3 small nucleolar RNA in providing an intertwined RNA-protein assembly platform for the separate maturation of 18S rRNA domains. The strategic placement of a molecular motor at the center of the particle further suggests a mechanism for mediating conformational changes within this giant particle. This study provides a structural framework for a mechanistic understanding of eukaryotic ribosome assembly in the model organism Saccharomyces cerevisiae.