Summary information and primary citation
- PDB-id
- 5u34; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-RNA
- Method
- X-ray (3.255 Å)
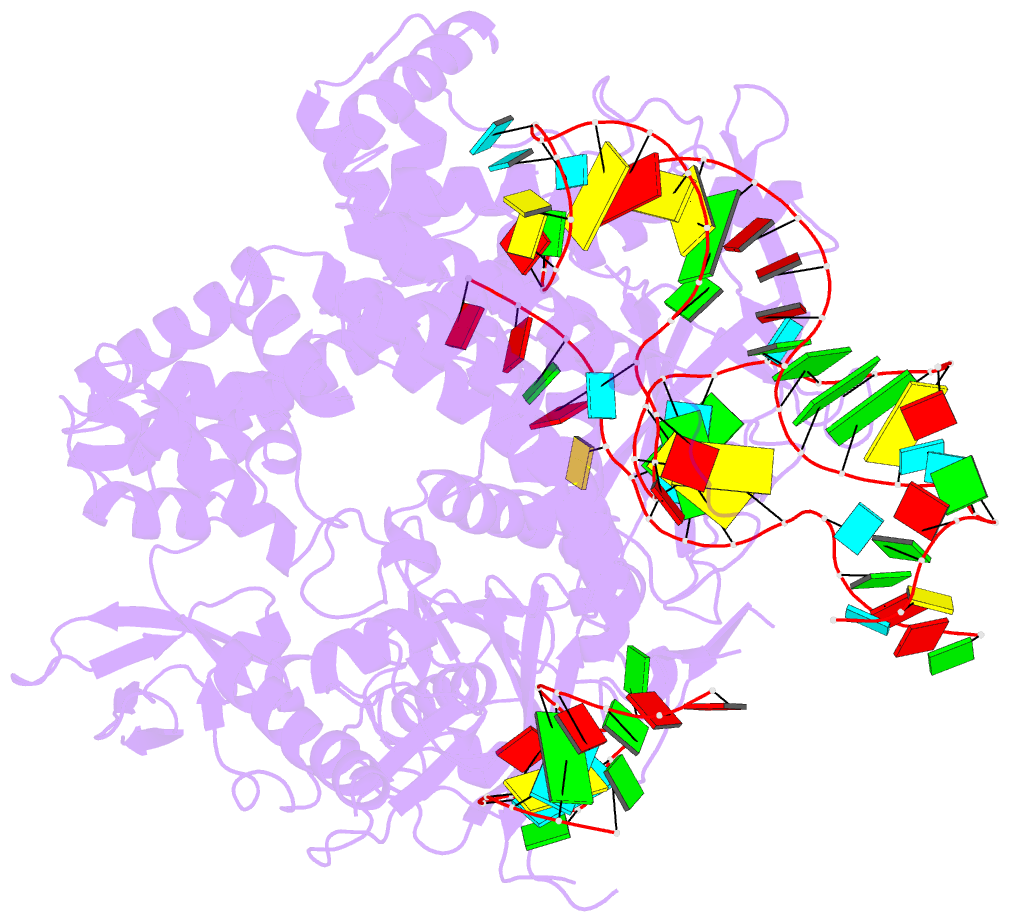
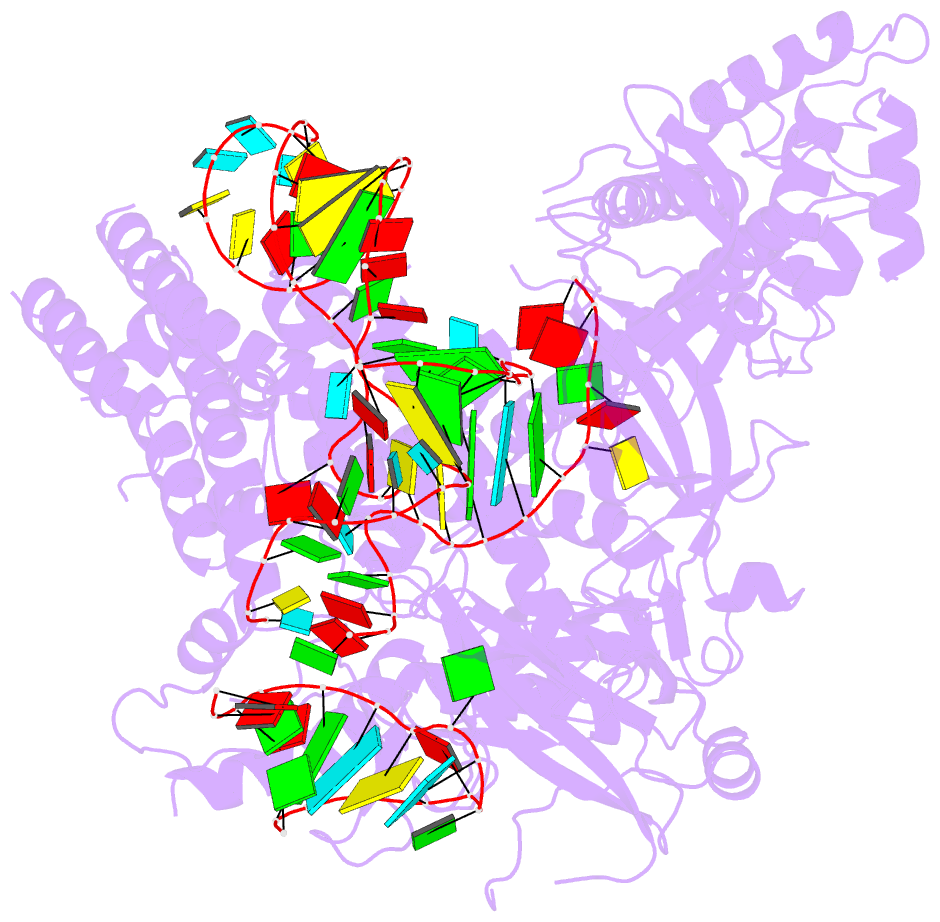
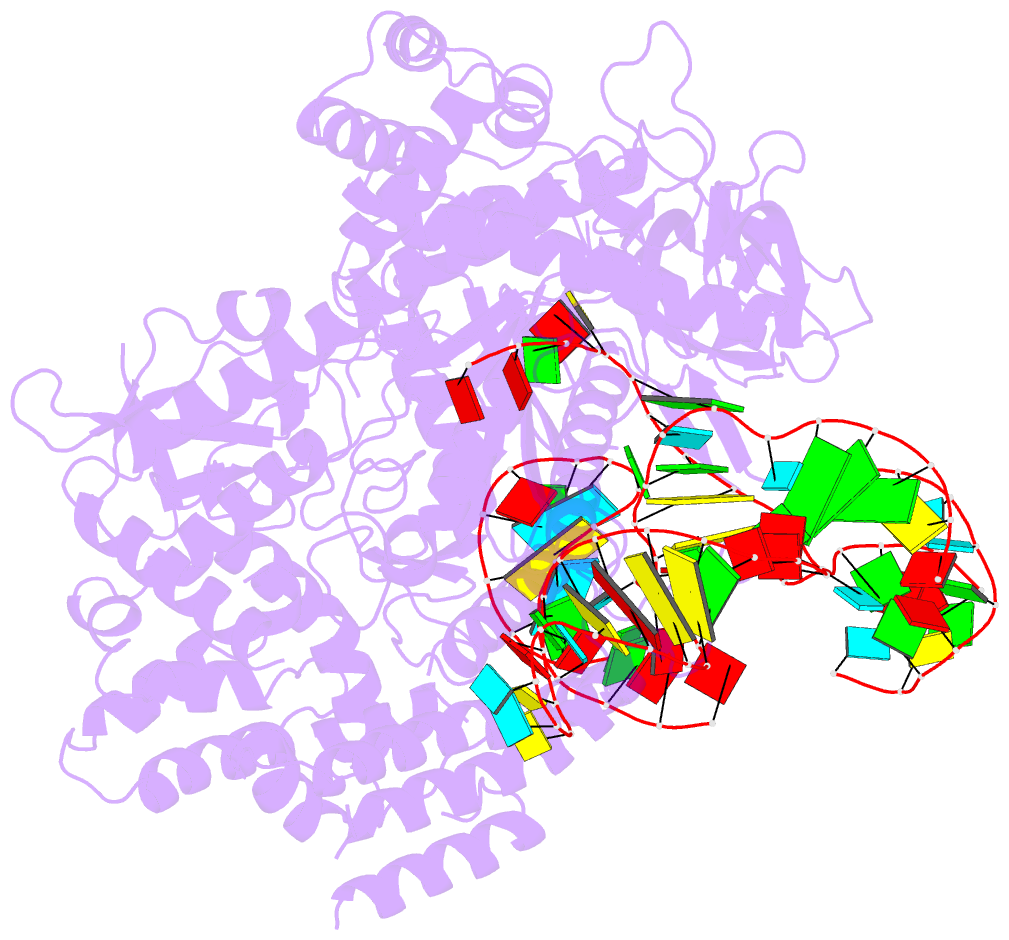
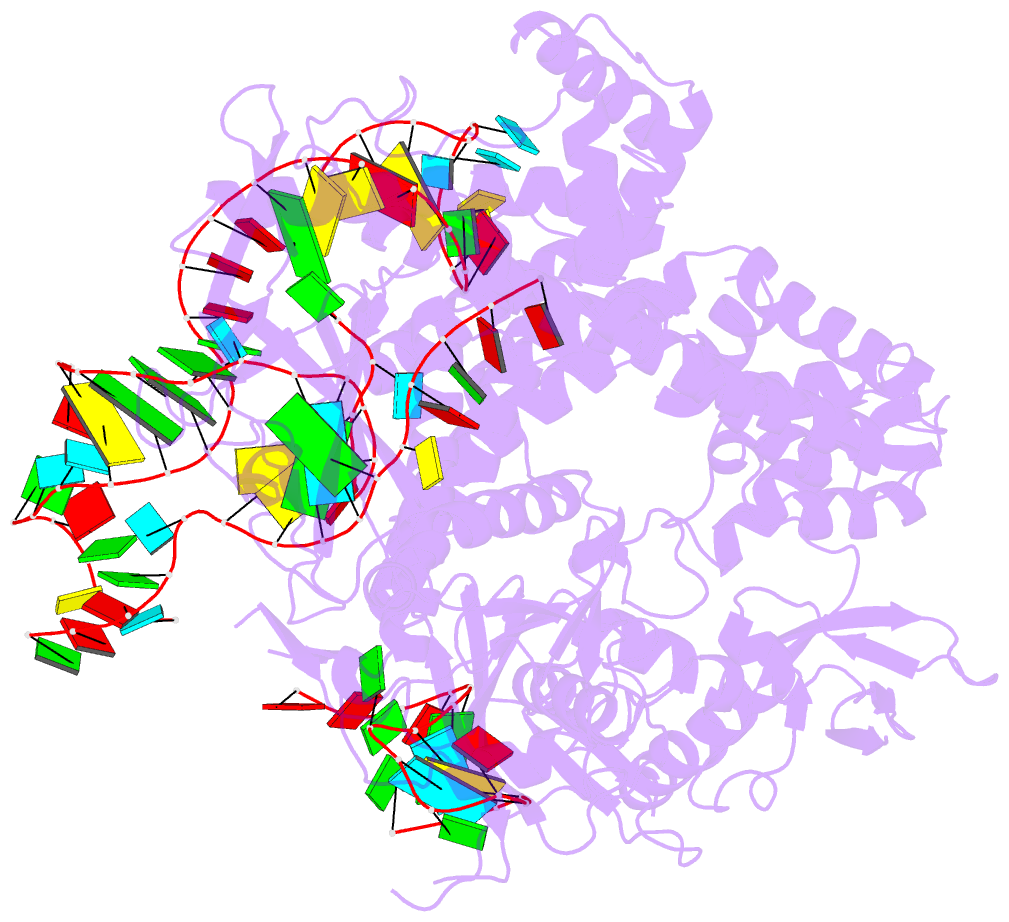
- Summary
- Crystal structure of aacc2c1-sgrna binary complex
- Reference
- Yang H, Gao P, Rajashankar KR, Patel DJ (2016): "PAM-Dependent Target DNA Recognition and Cleavage by C2c1 CRISPR-Cas Endonuclease." Cell, 167, 1814-1828.e12. doi: 10.1016/j.cell.2016.11.053.
- Abstract
- C2c1 is a newly identified guide RNA-mediated type V-B CRISPR-Cas endonuclease that site-specifically targets and cleaves both strands of target DNA. We have determined crystal structures of Alicyclobacillus acidoterrestris C2c1 (AacC2c1) bound to sgRNA as a binary complex and to target DNAs as ternary complexes, thereby capturing catalytically competent conformations of AacC2c1 with both target and non-target DNA strands independently positioned within a single RuvC catalytic pocket. Moreover, C2c1-mediated cleavage results in a staggered seven-nucleotide break of target DNA. crRNA adopts a pre-ordered five-nucleotide A-form seed sequence in the binary complex, with release of an inserted tryptophan, facilitating zippering up of 20-bp guide RNA:target DNA heteroduplex on ternary complex formation. Notably, the PAM-interacting cleft adopts a "locked" conformation on ternary complex formation. Structural comparison of C2c1 ternary complexes with their Cas9 and Cpf1 counterparts highlights the diverse mechanisms adopted by these distinct CRISPR-Cas systems, thereby broadening and enhancing their applicability as genome editing tools.