Summary information and primary citation
- PDB-id
- 5u91; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- isomerase-DNA
- Method
- X-ray (3.104 Å)
- Summary
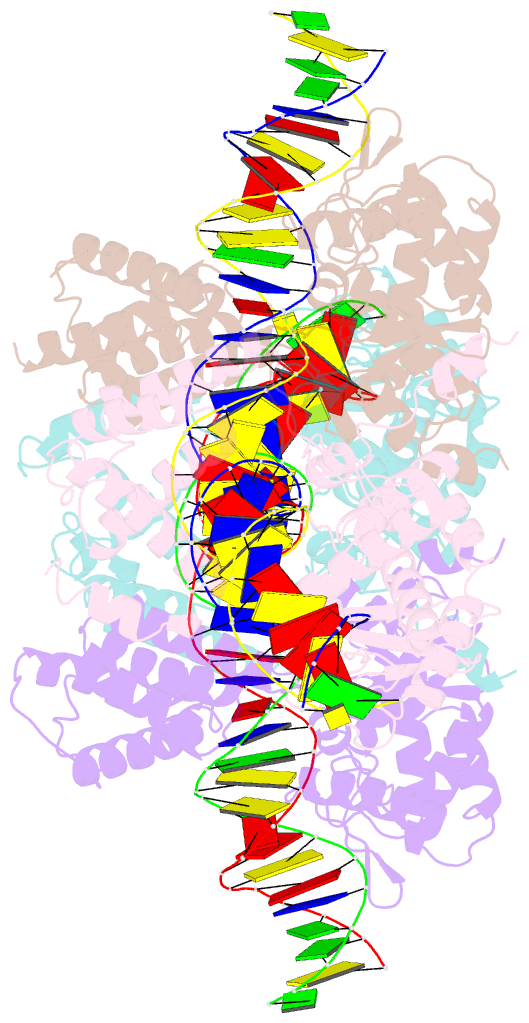
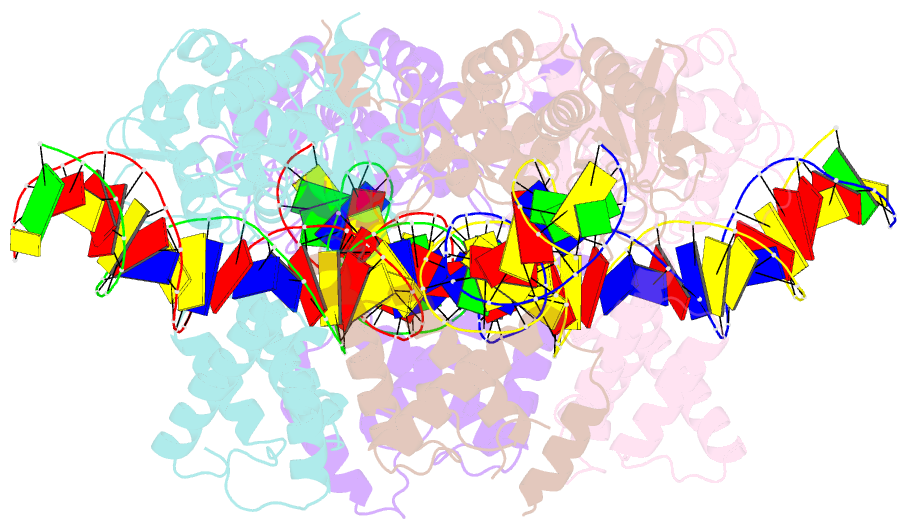
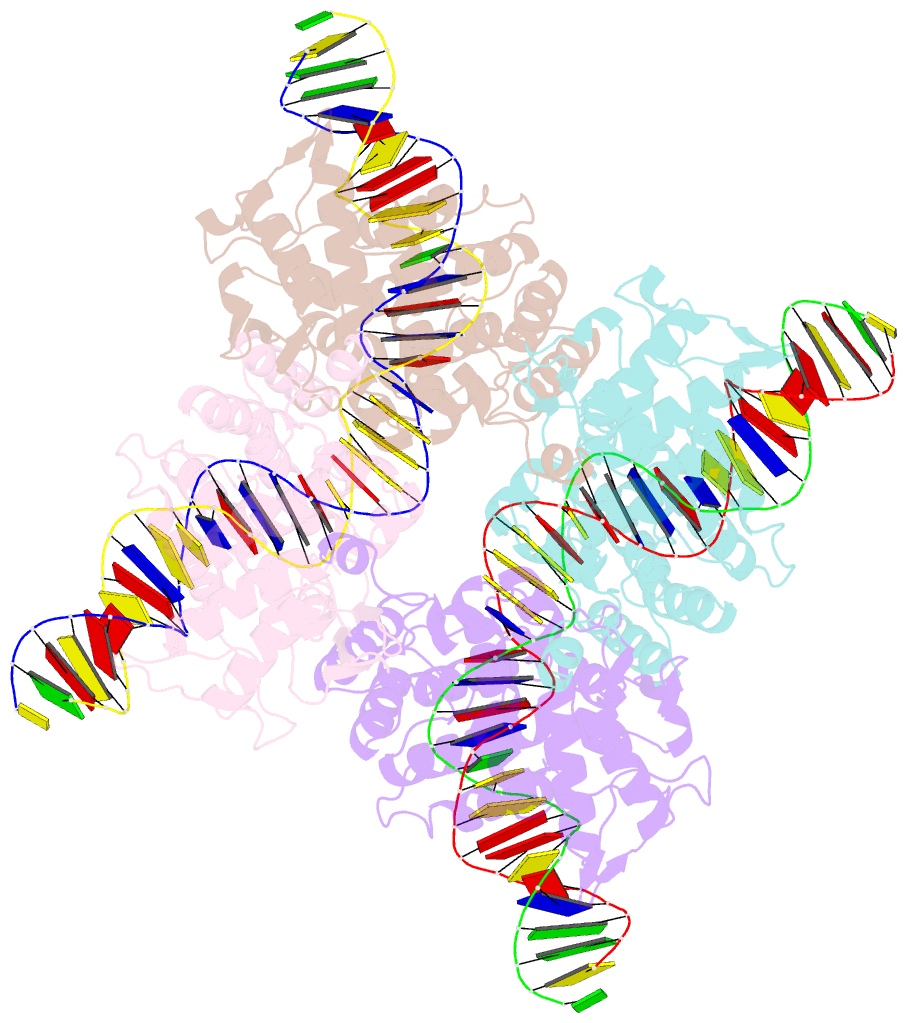
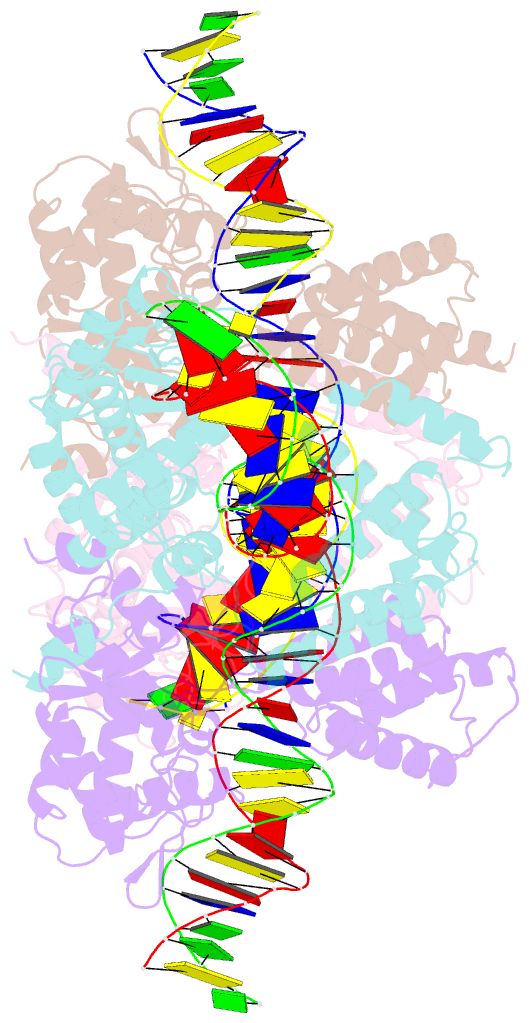
- Crystal structure of tre-loxltr complex
- Reference
- Meinke G, Karpinski J, Buchholz F, Bohm A (2017): "Crystal structure of an engineered, HIV-specific recombinase for removal of integrated proviral DNA." Nucleic Acids Res., 45, 9726-9740. doi: 10.1093/nar/gkx603.
- Abstract
- As part of the HIV infection cycle, viral DNA inserts into the genome of host cells such that the integrated DNA encoding the viral proteins is flanked by long terminal repeat (LTR) regions from the retrovirus. In an effort to develop novel genome editing techniques that safely excise HIV provirus from cells, Tre, an engineered version of Cre recombinase, was designed to target a 34-bp sequence within the HIV-1 LTR (loxLTR). The sequence targeted by Tre lacks the symmetry present in loxP, the natural DNA substrate for Cre. We report here the crystal structure of a catalytically inactive (Y324F) mutant of this engineered Tre recombinase in complex with the loxLTR DNA substrate. We also report that 17 of the 19 amino acid changes relative to Cre contribute to the altered specificity, even though many of these residues do not contact the DNA directly. We hypothesize that some mutations increase the flexibility of the Cre tetramer and that this, along with flexibility in the DNA, enable the engineered enzyme and DNA substrate to adopt complementary conformations.