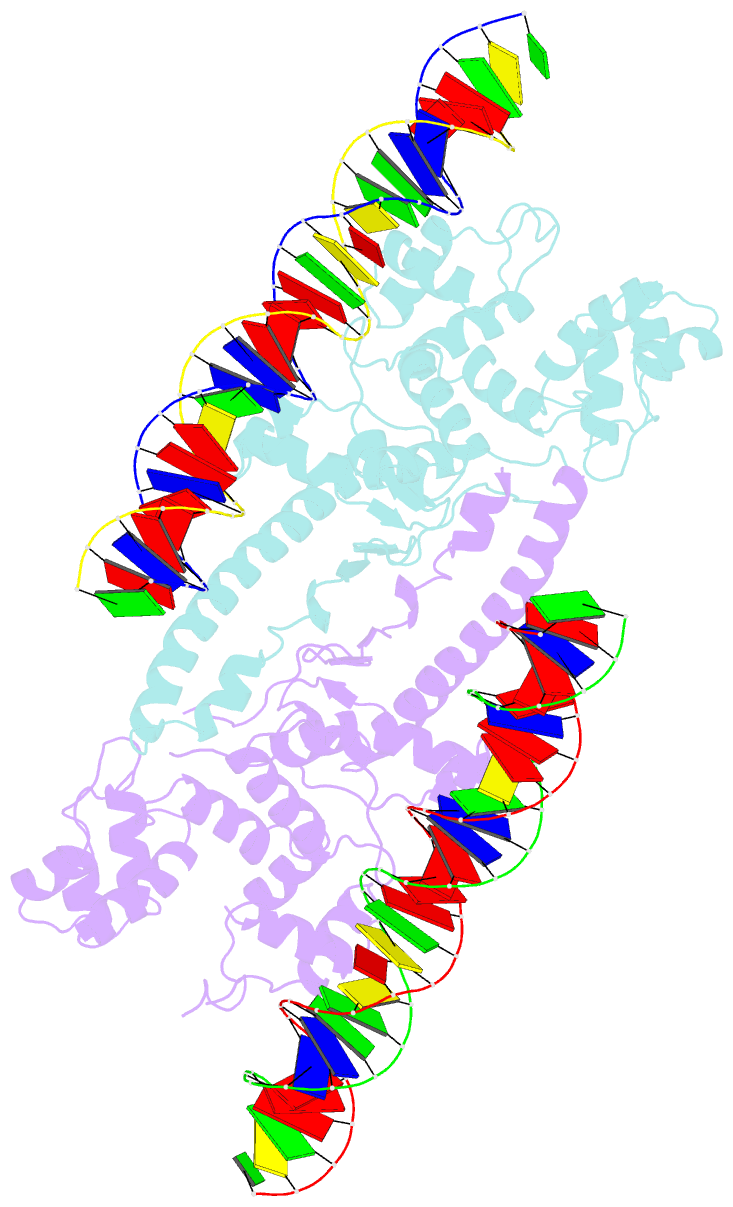
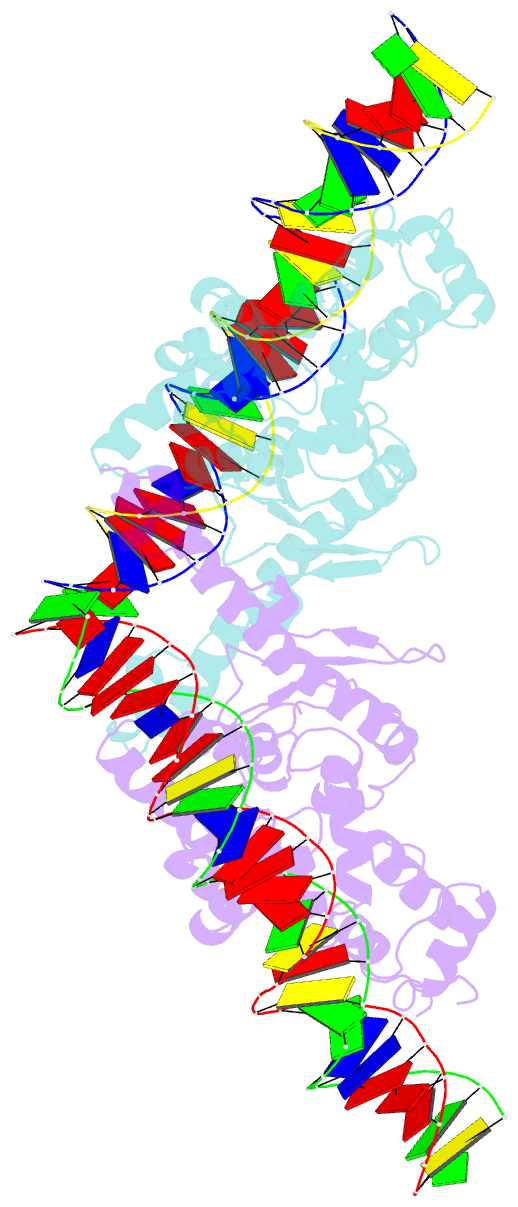
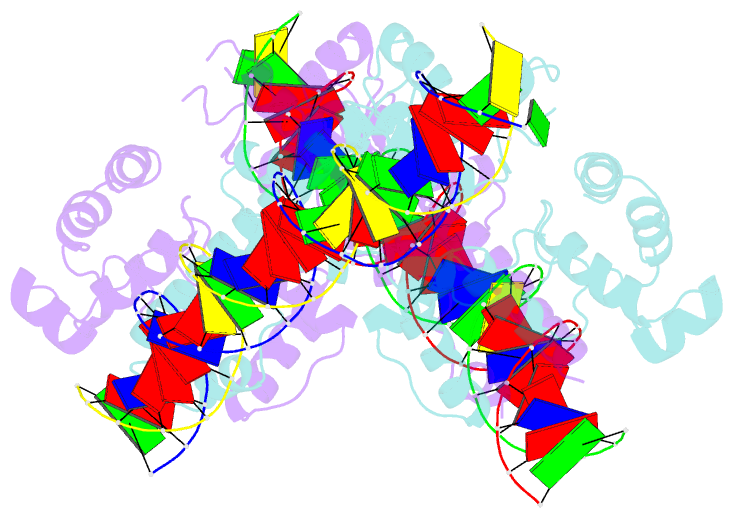
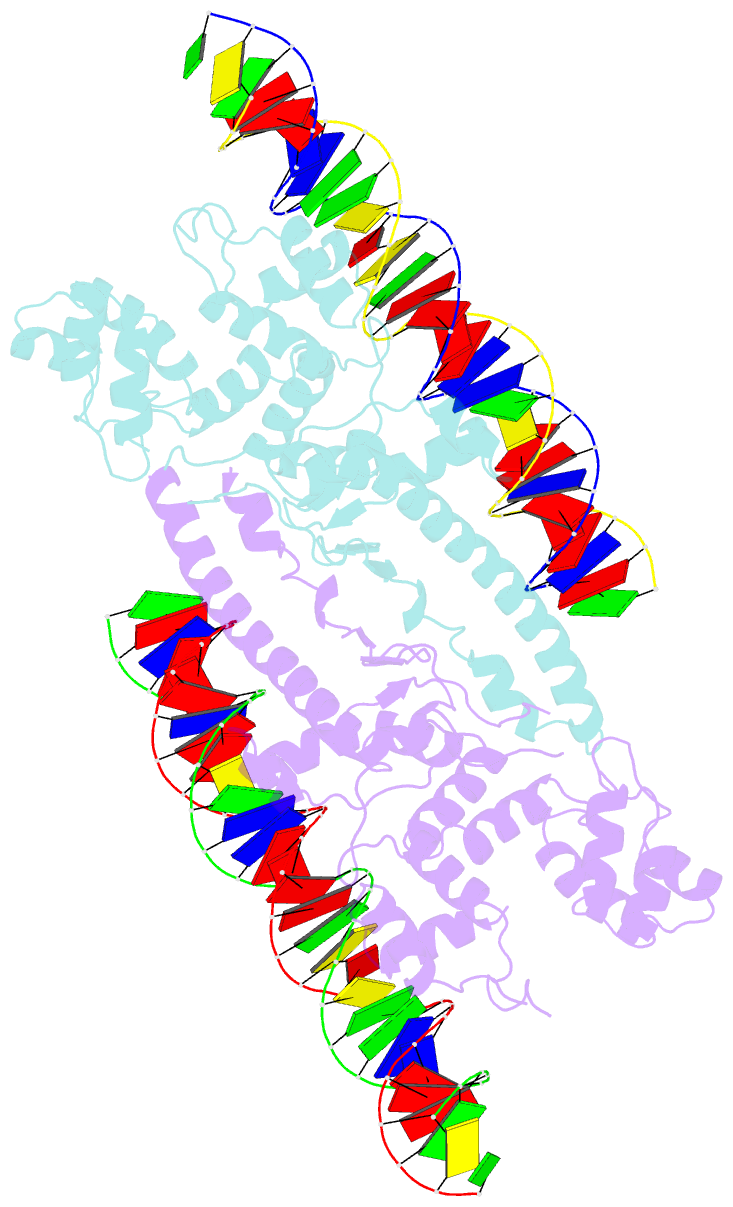
Summary information and primary citation
- PDB-id
- 5ui5; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (3.4 Å)
- Summary
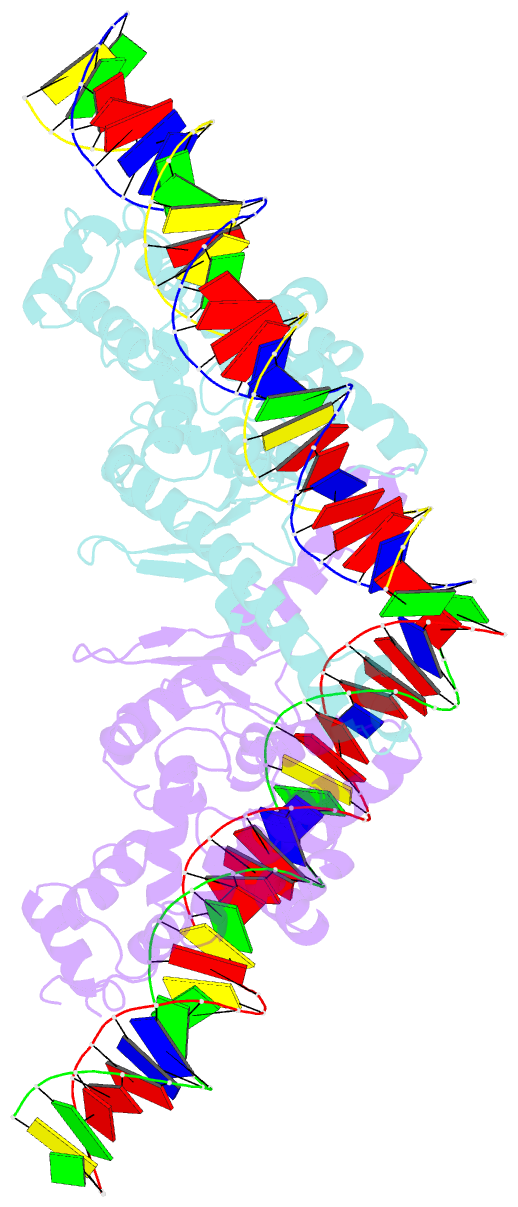
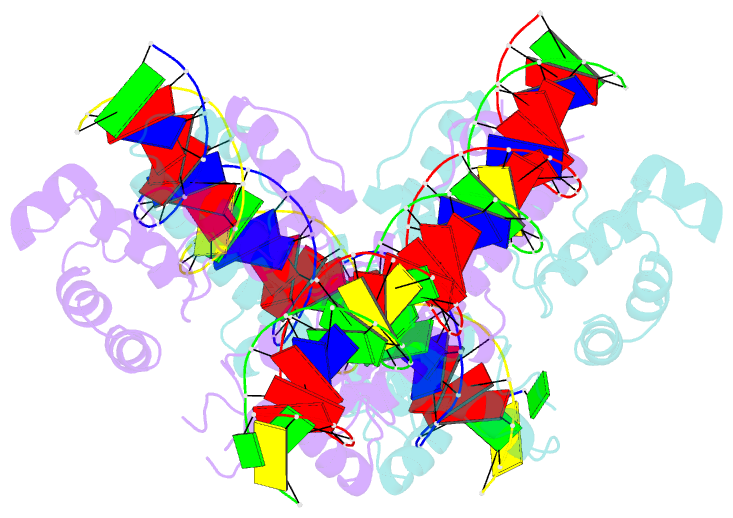
- Crystal structure of aquifex aeolicus sigman bound to promoter DNA
- Reference
- Campbell EA, Kamath S, Rajashankar KR, Wu M, Darst SA (2017): "Crystal structure of Aquifex aeolicus sigma (N) bound to promoter DNA and the structure of sigma (N)-holoenzyme." Proc. Natl. Acad. Sci. U.S.A., 114, E1805-E1814. doi: 10.1073/pnas.1619464114.
- Abstract
- The bacterial σ factors confer promoter specificity to the RNA polymerase (RNAP). One alternative σ factor, σN, is unique in its structure and functional mechanism, forming transcriptionally inactive promoter complexes that require activation by specialized AAA+ ATPases. We report a 3.4-Å resolution X-ray crystal structure of a σN fragment in complex with its cognate promoter DNA, revealing the molecular details of promoter recognition by σN The structure allowed us to build and refine an improved σN-holoenzyme model based on previously published 3.8-Å resolution X-ray data. The improved σN-holoenzyme model reveals a conserved interdomain interface within σN that, when disrupted by mutations, leads to transcription activity without activator intervention (so-called bypass mutants). Thus, the structure and stability of this interdomain interface are crucial for the role of σN in blocking transcription activity and in maintaining the activator sensitivity of σN.