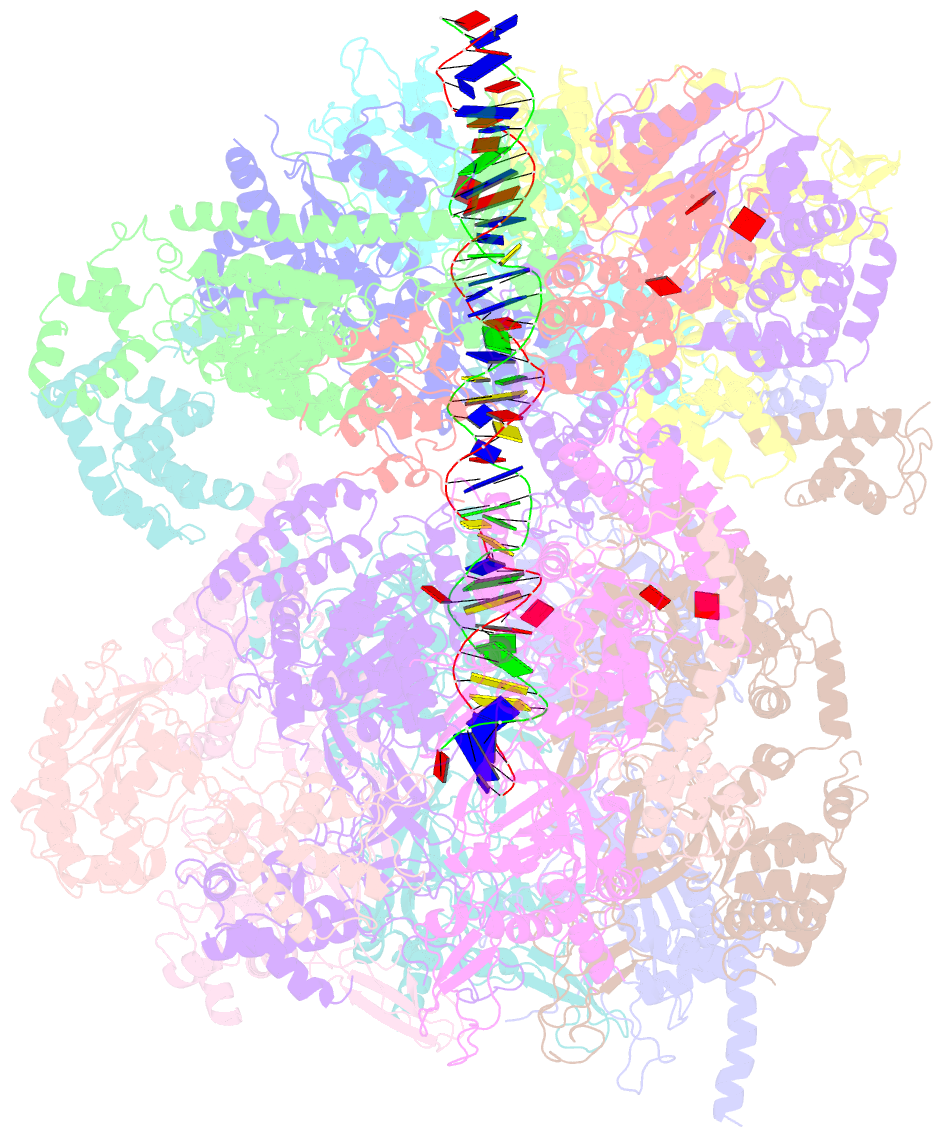
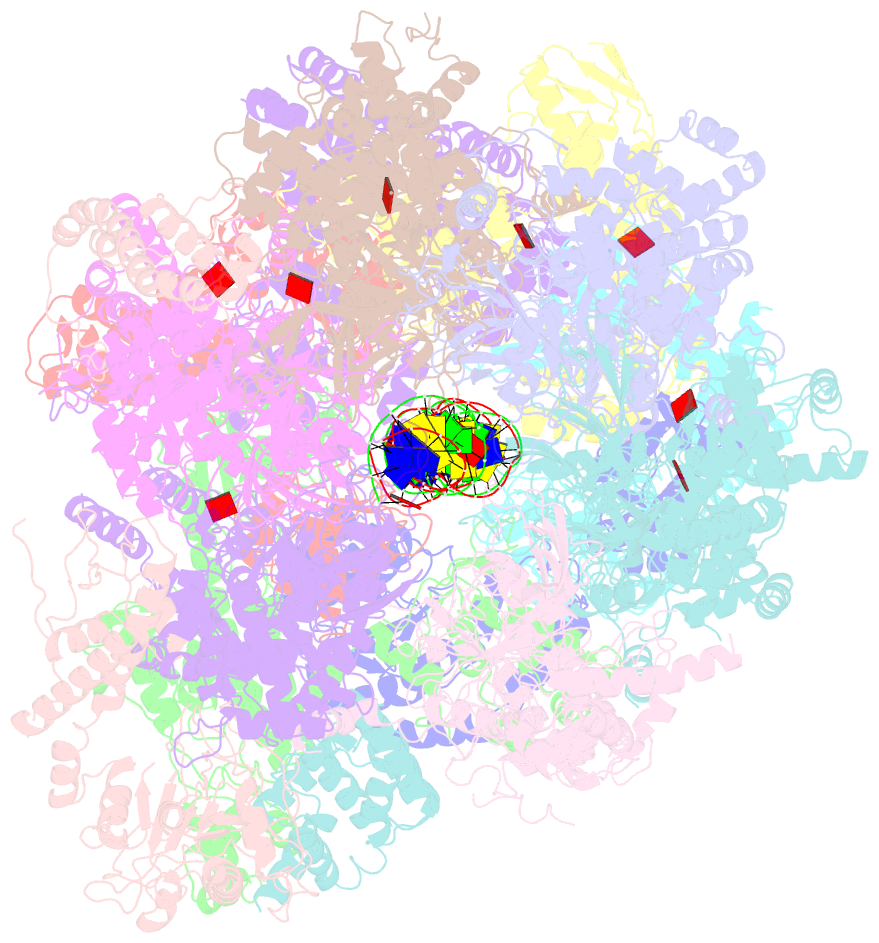
Summary information and primary citation
- PDB-id
- 5v8f; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- replication
- Method
- cryo-EM (3.9 Å)
- Summary
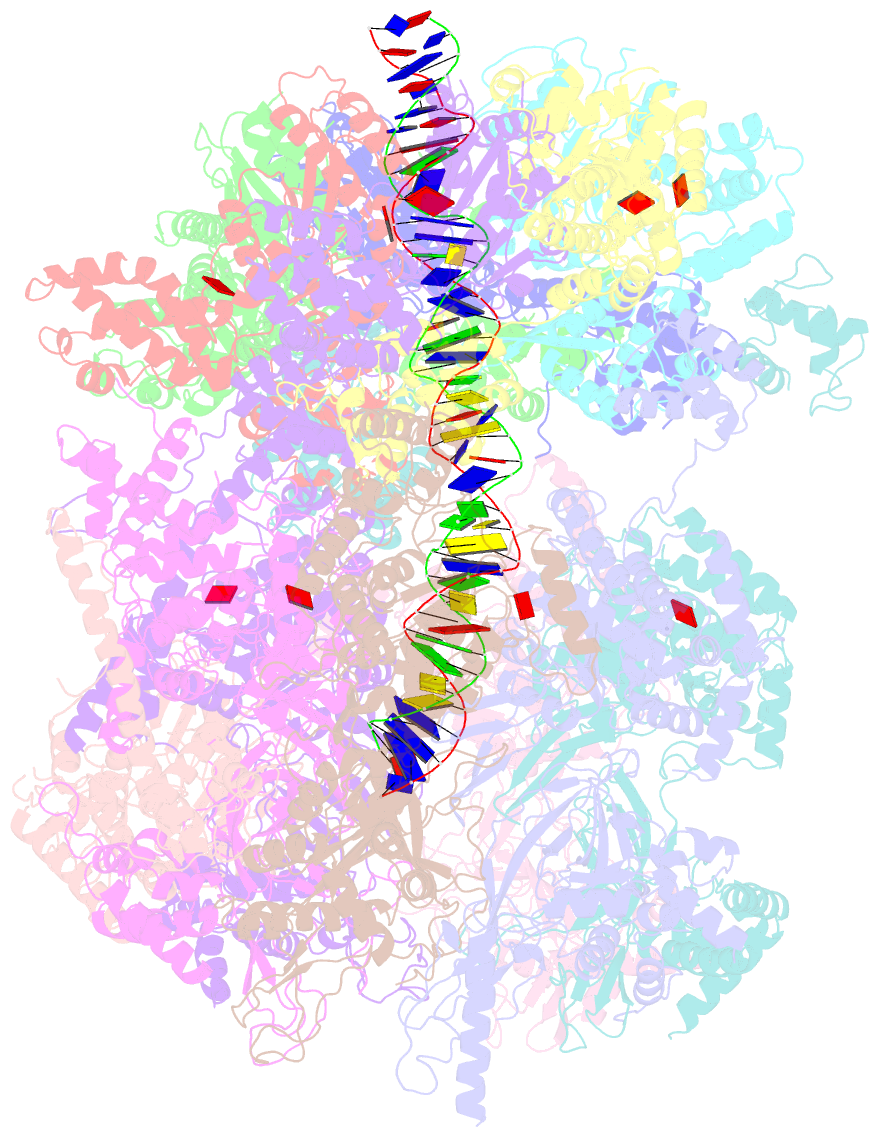
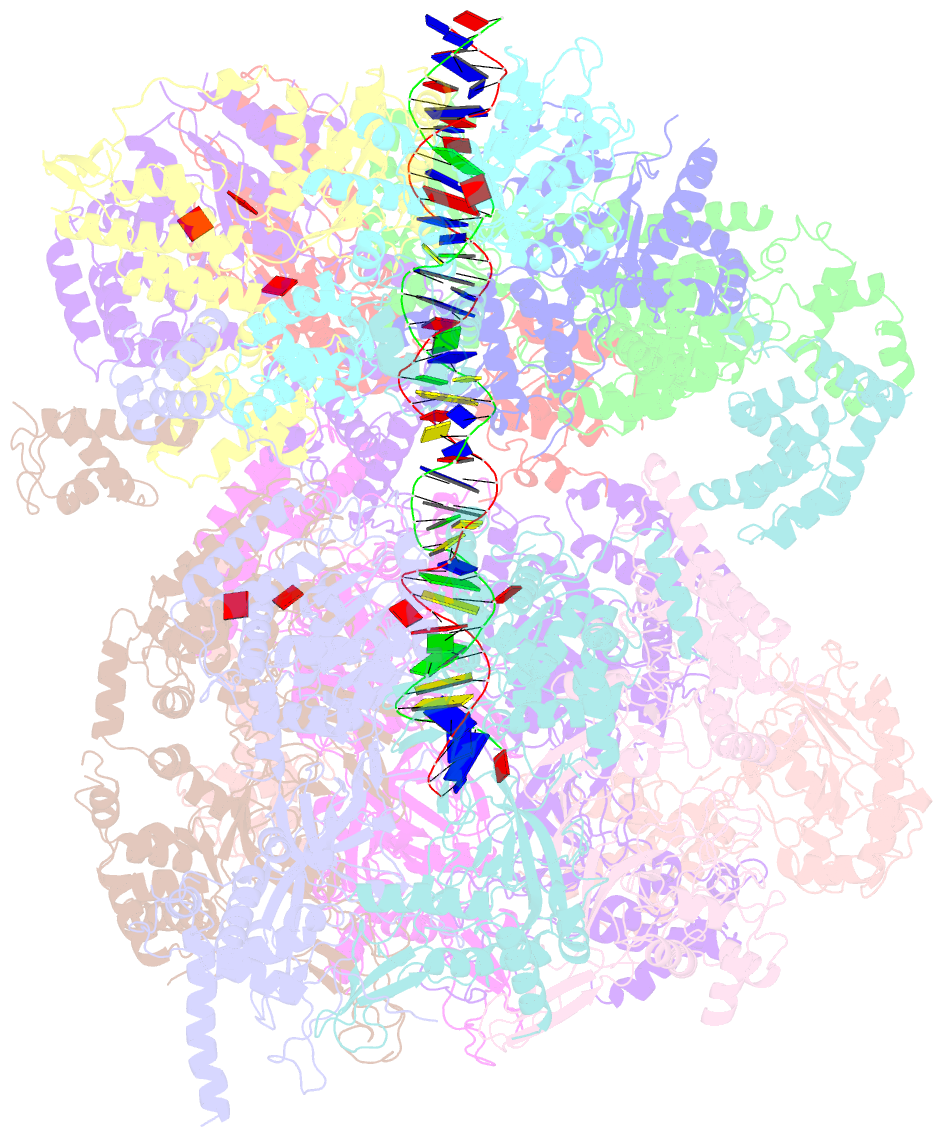
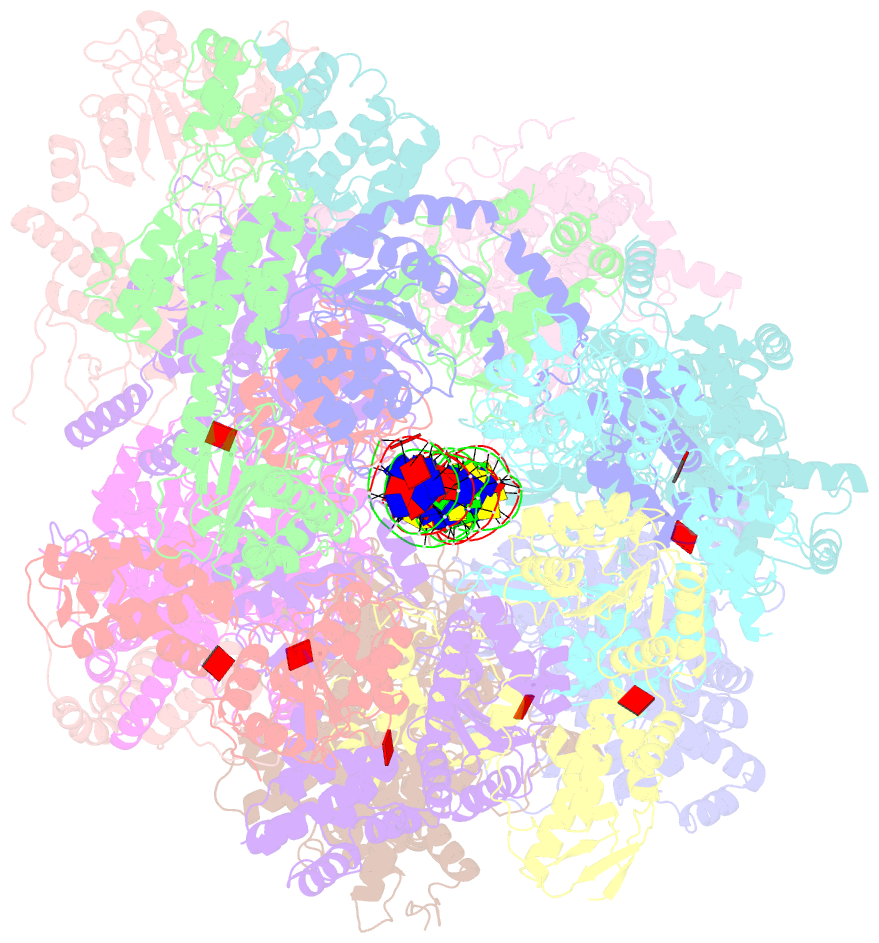
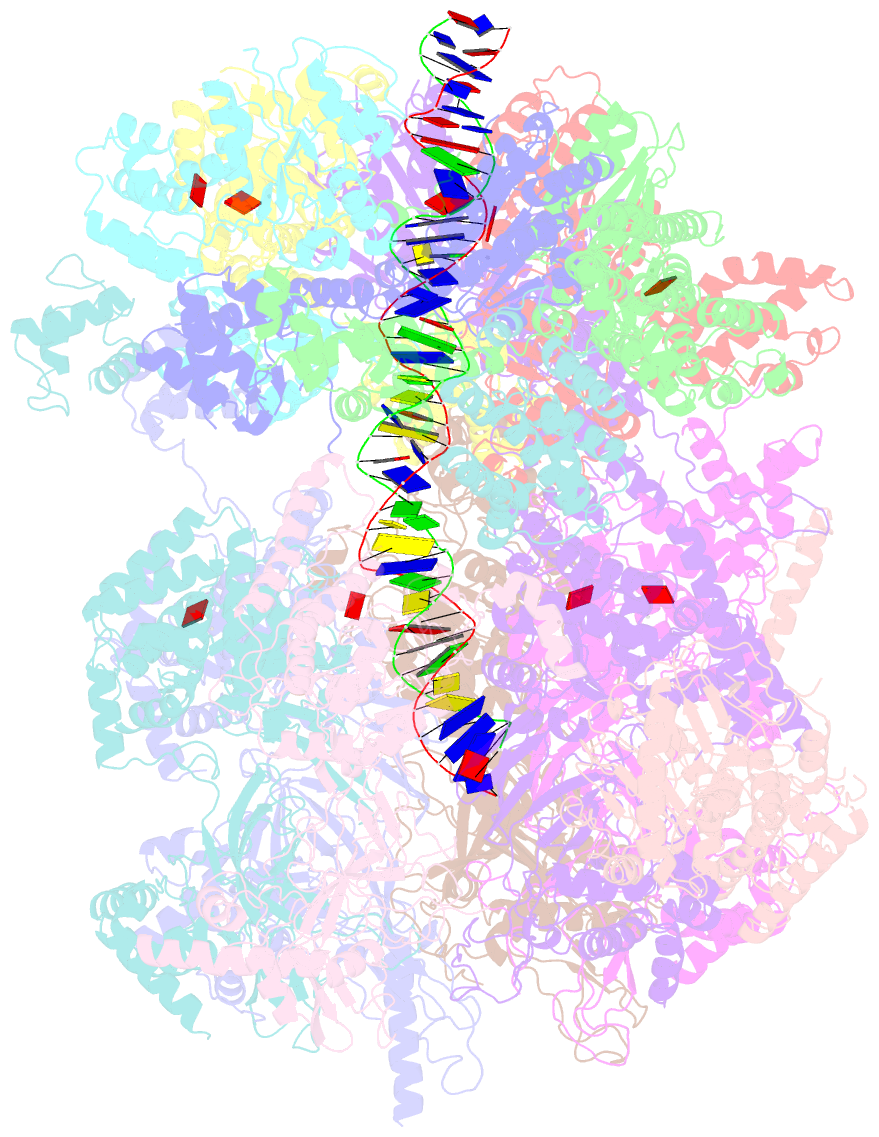
- Structural basis of mcm2-7 replicative helicase loading by orc-cdc6 and cdt1
- Reference
- Yuan Z, Riera A, Bai L, Sun J, Nandi S, Spanos C, Chen ZA, Barbon M, Rappsilber J, Stillman B, Speck C, Li H (2017): "Structural basis of Mcm2-7 replicative helicase loading by ORC-Cdc6 and Cdt1." Nat. Struct. Mol. Biol., 24, 316-324. doi: 10.1038/nsmb.3372.
- Abstract
- To initiate DNA replication, the origin recognition complex (ORC) and Cdc6 load an Mcm2-7 double hexamer onto DNA. Without ATP hydrolysis, ORC-Cdc6 recruits one Cdt1-bound Mcm2-7 hexamer, thus forming an ORC-Cdc6-Cdt1-Mcm2-7 (OCCM) helicase-loading intermediate. Here we report a 3.9-Å structure of Saccharomyces cerevisiae OCCM on DNA. Flexible Mcm2-7 winged-helix domains (WHDs) engage ORC-Cdc6. A three-domain Cdt1 configuration embraces Mcm2, Mcm4, and Mcm6, thus comprising nearly half of the hexamer. The Cdt1 C-terminal domain extends to the Mcm6 WHD, which binds the Orc4 WHD. DNA passes through the ORC-Cdc6 and Mcm2-7 rings. Origin DNA interaction is mediated by an α-helix within Orc4 and positively charged loops within Orc2 and Cdc6. The Mcm2-7 C-tier AAA+ ring is topologically closed by an Mcm5 loop that embraces Mcm2, but the N-tier-ring Mcm2-Mcm5 interface remains open. This structure suggests a loading mechanism of the first Cdt1-bound Mcm2-7 hexamer by ORC-Cdc6.