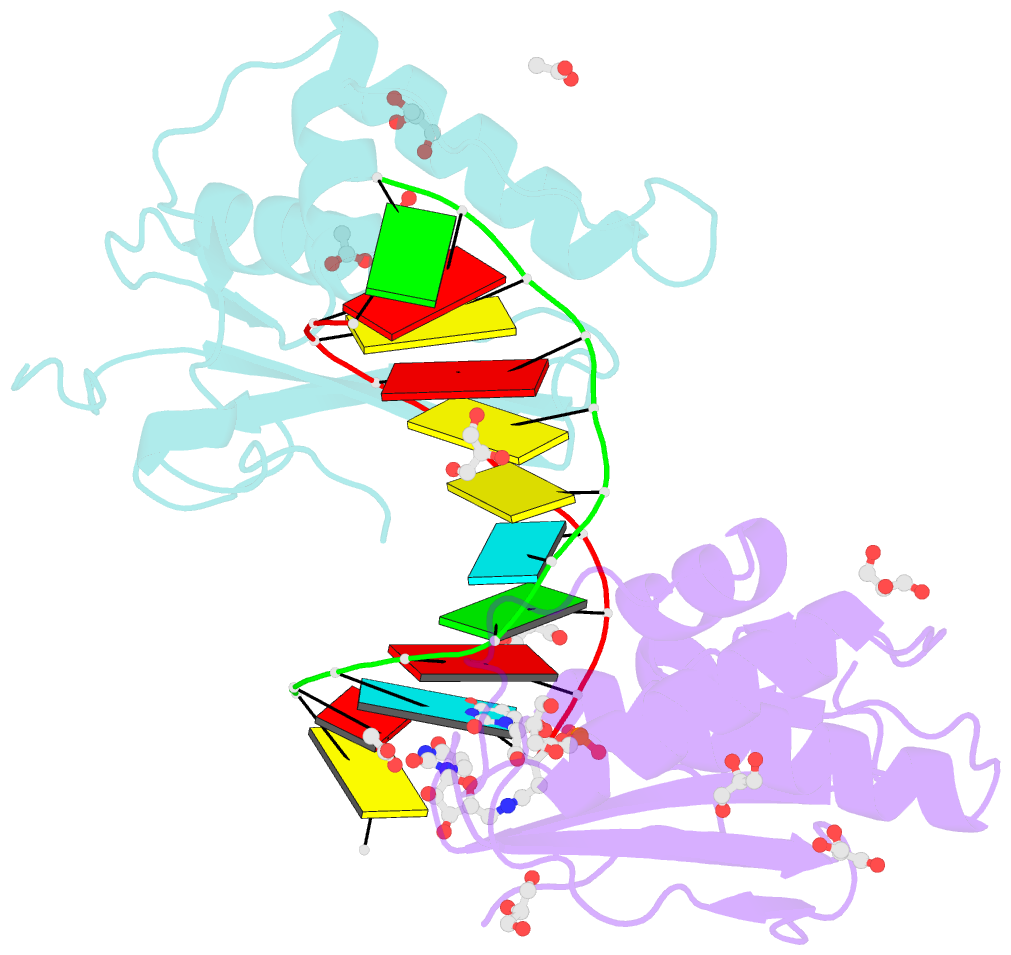
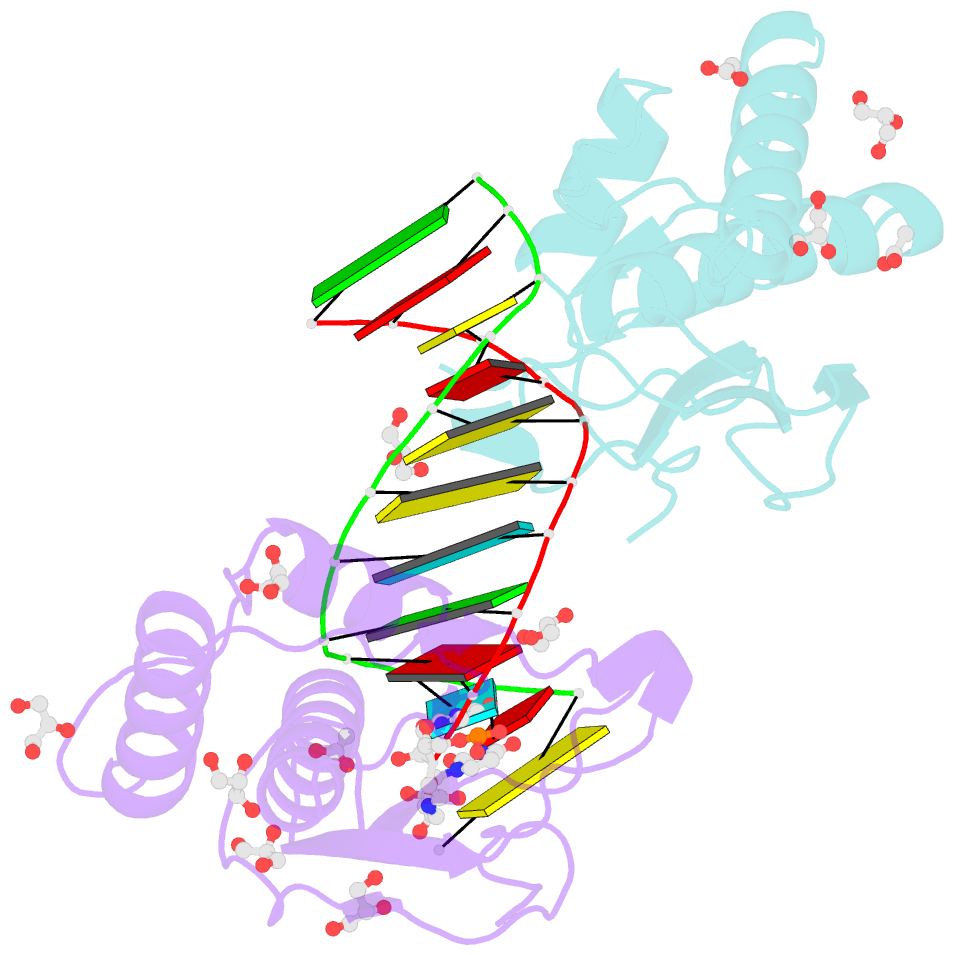
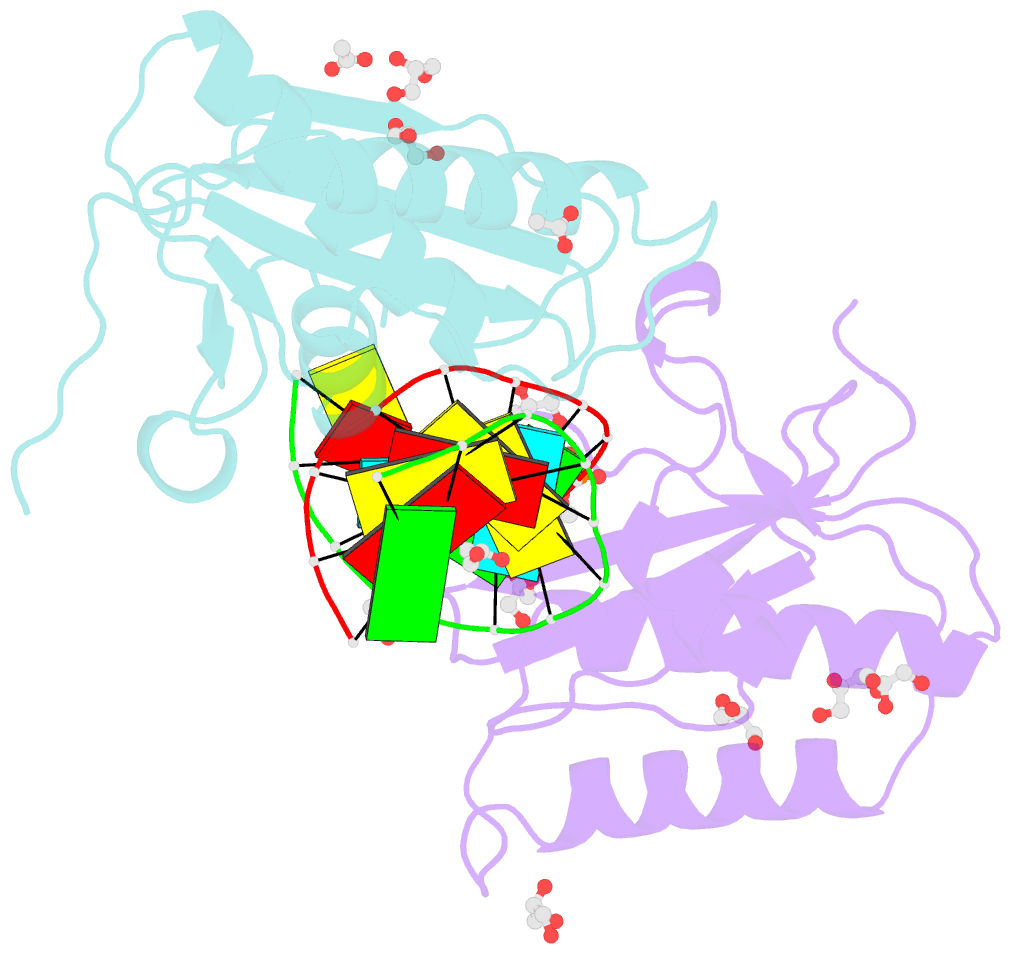
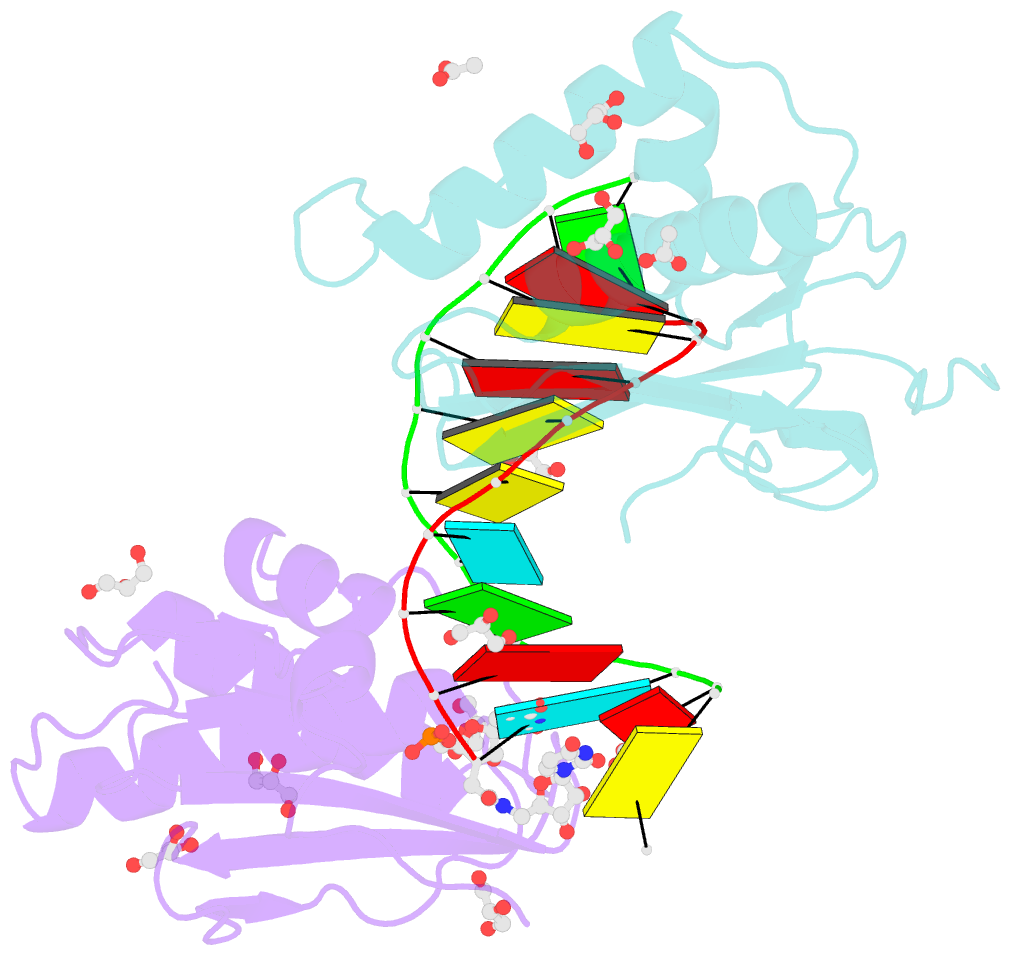
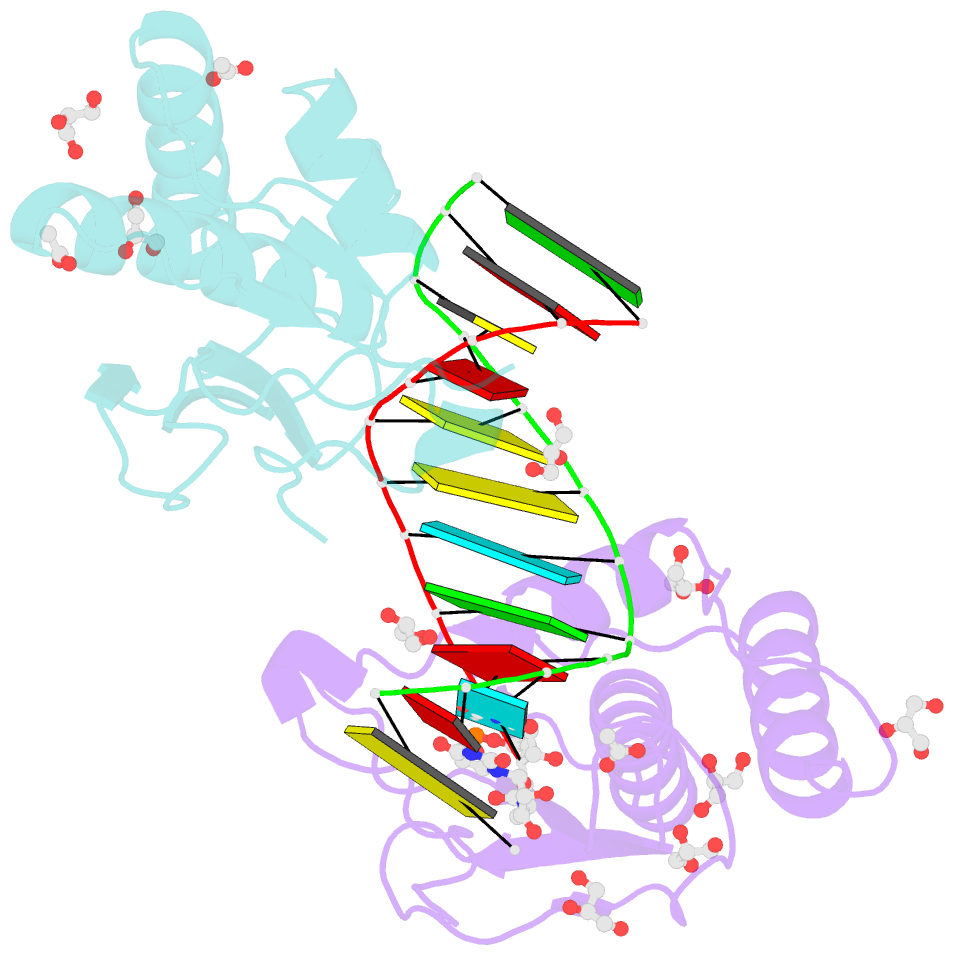
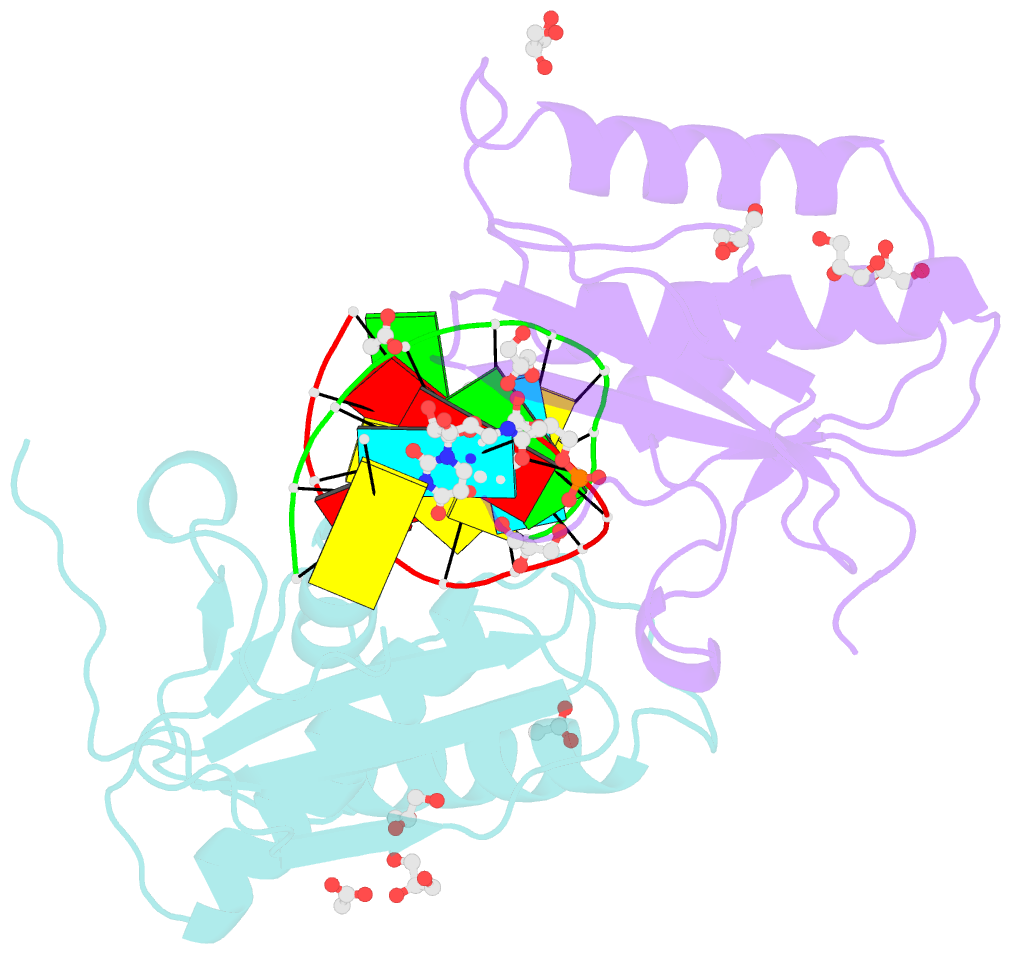
Summary information and primary citation
- PDB-id
- 5vaj; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-RNA-DNA
- Method
- X-ray (1.95 Å)
- Summary
- Bhrnase h - amide-RNA-DNA complex
- Reference
- Mutisya D, Hardcastle T, Cheruiyot SK, Pallan PS, Kennedy SD, Egli M, Kelley ML, Smith AVB, Rozners E (2017): "Amide linkages mimic phosphates in RNA interactions with proteins and are well tolerated in the guide strand of short interfering RNAs." Nucleic Acids Res., 45, 8142-8155. doi: 10.1093/nar/gkx558.
- Abstract
- While the use of RNA interference (RNAi) in molecular biology and functional genomics is a well-established technology, in vivo applications of synthetic short interfering RNAs (siRNAs) require chemical modifications. We recently found that amides as non-ionic replacements for phosphodiesters may be useful modifications for optimization of siRNAs. Herein, we report a comprehensive study of systematic replacement of a single phosphate with an amide linkage throughout the guide strand of siRNAs. The results show that amides are surprisingly well tolerated in the seed and central regions of the guide strand and increase the silencing activity when placed between nucleosides 10 and 12, at the catalytic site of Argonaute. A potential explanation is provided by the first crystal structure of an amide-modified RNA-DNA with Bacillus halodurans RNase H1. The structure reveals how small changes in both RNA and protein conformation allow the amide to establish hydrogen bonding interactions with the protein. Molecular dynamics simulations suggest that these alternative binding modes may compensate for interactions lost due to the absence of a phosphodiester moiety. Our results suggest that an amide can mimic important hydrogen bonding interactions with proteins required for RNAi activity and may be a promising modification for optimization of biological properties of siRNAs.