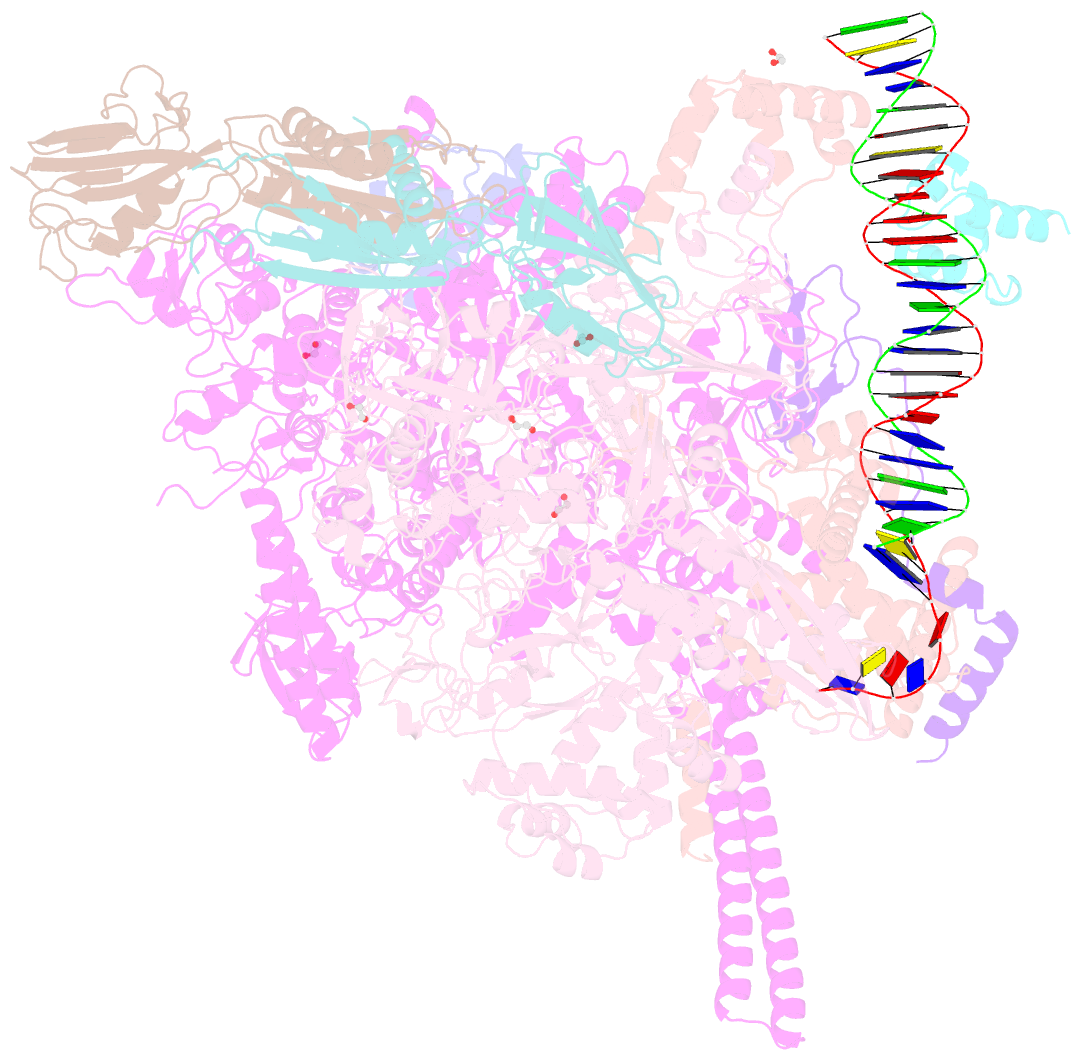
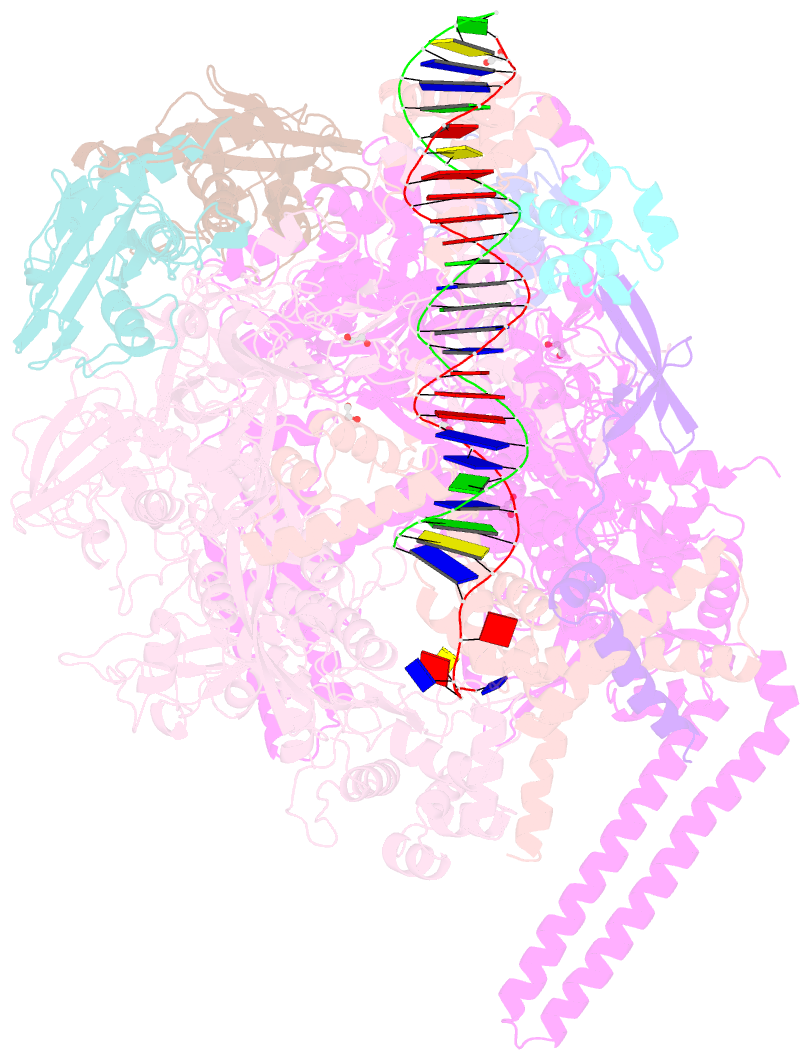
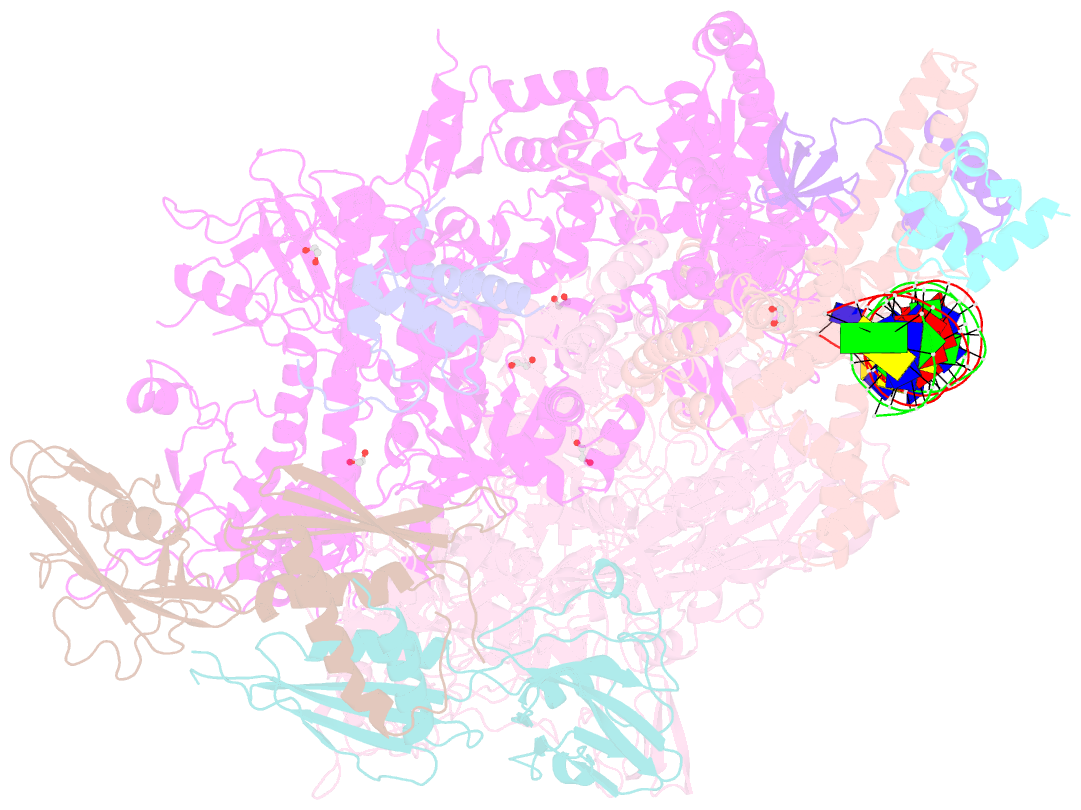
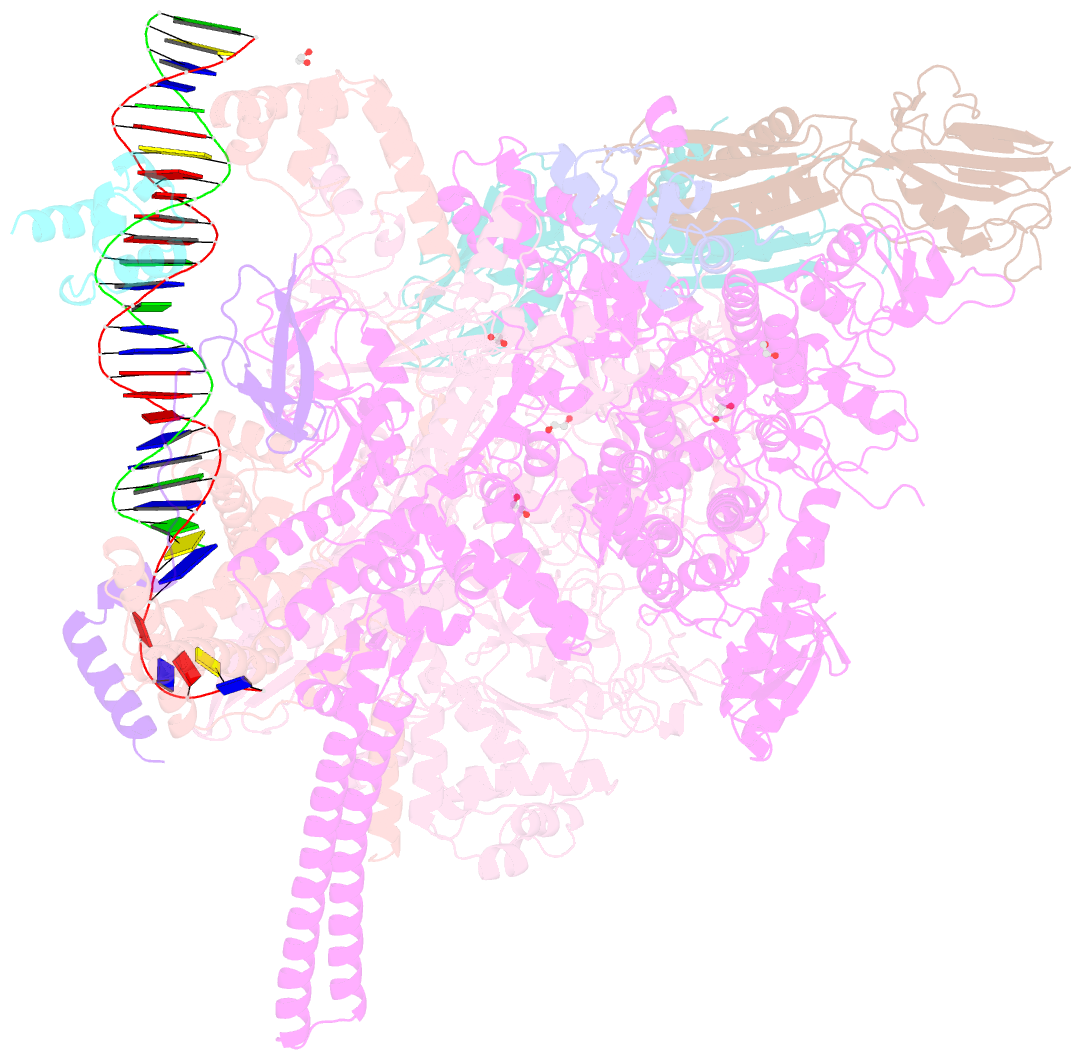
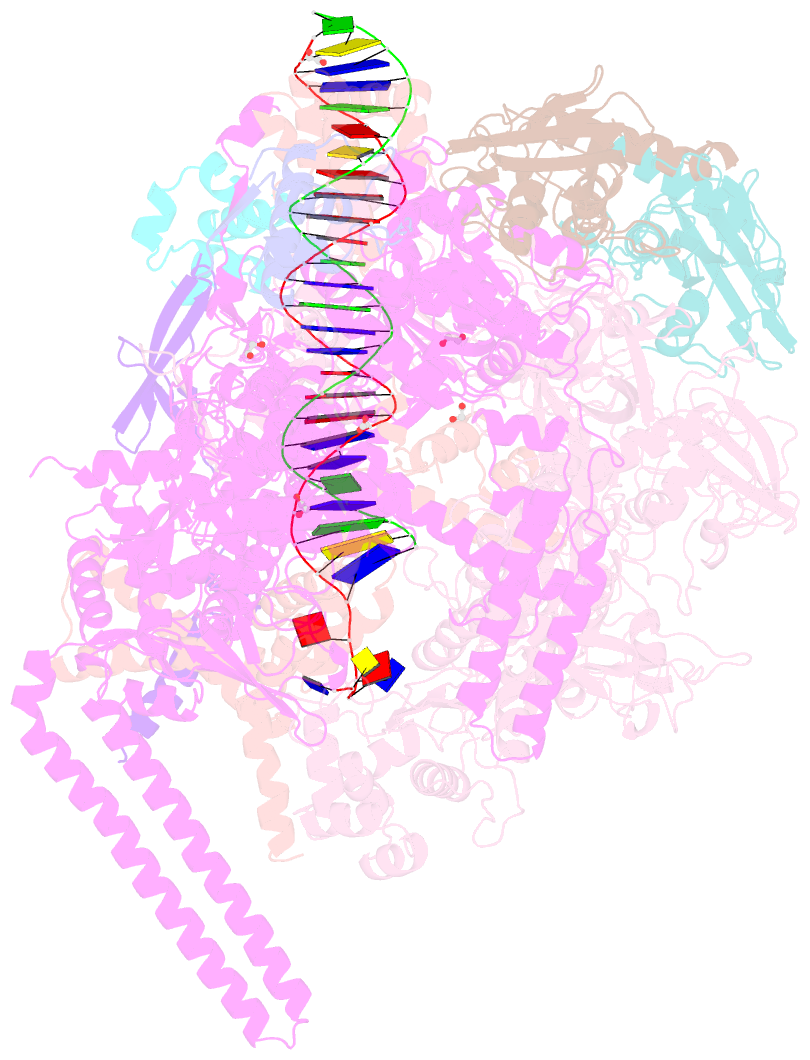
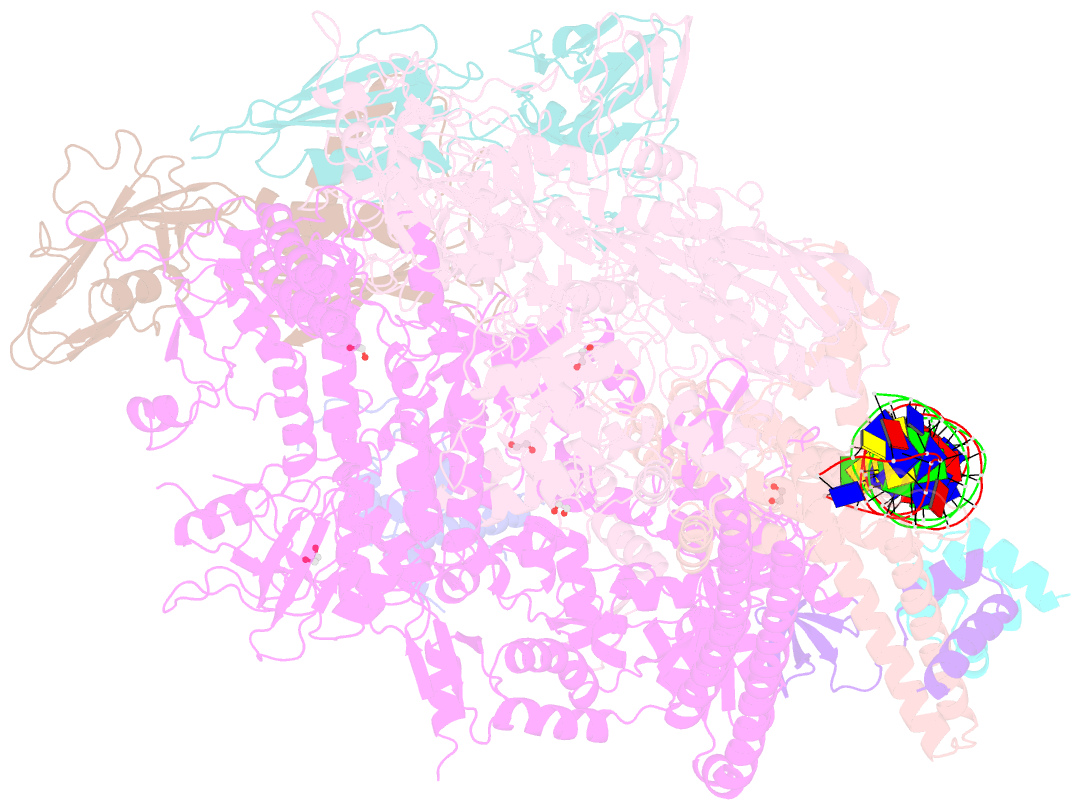
Summary information and primary citation
- PDB-id
- 5vi8; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- X-ray (2.76 Å)
- Summary
- Structure of a mycobacterium smegmatis transcription initiation complex with an upstream-fork promoter fragment
- Reference
- Hubin EA, Lilic M, Darst SA, Campbell EA (2017): "Structural insights into the mycobacteria transcription initiation complex from analysis of X-ray crystal structures." Nat Commun, 8, 16072. doi: 10.1038/ncomms16072.
- Abstract
- The mycobacteria RNA polymerase (RNAP) is a target for antimicrobials against tuberculosis, motivating structure/function studies. Here we report a 3.2 Å-resolution crystal structure of a Mycobacterium smegmatis (Msm) open promoter complex (RPo), along with structural analysis of the Msm RPo and a previously reported 2.76 Å-resolution crystal structure of an Msm transcription initiation complex with a promoter DNA fragment. We observe the interaction of the Msm RNAP α-subunit C-terminal domain (αCTD) with DNA, and we provide evidence that the αCTD may play a role in Mtb transcription regulation. Our results reveal the structure of an Actinobacteria-unique insert of the RNAP β' subunit. Finally, our analysis reveals the disposition of the N-terminal segment of Msm σA, which may comprise an intrinsically disordered protein domain unique to mycobacteria. The clade-specific features of the mycobacteria RNAP provide clues to the profound instability of mycobacteria RPo compared with E. coli.