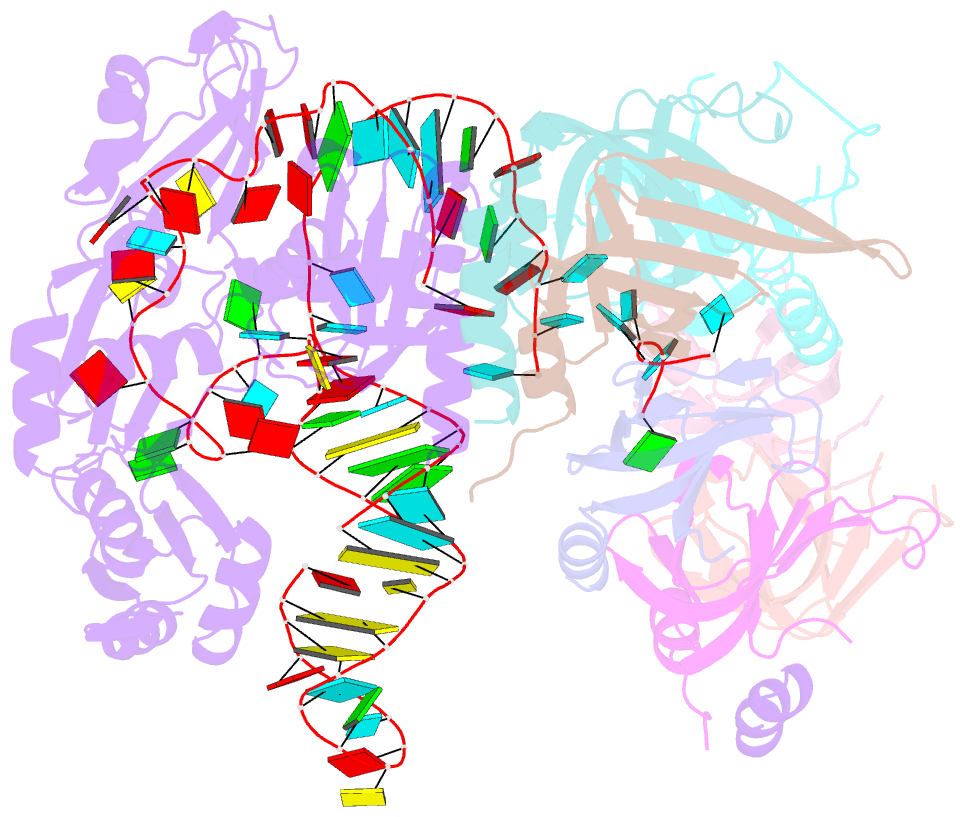
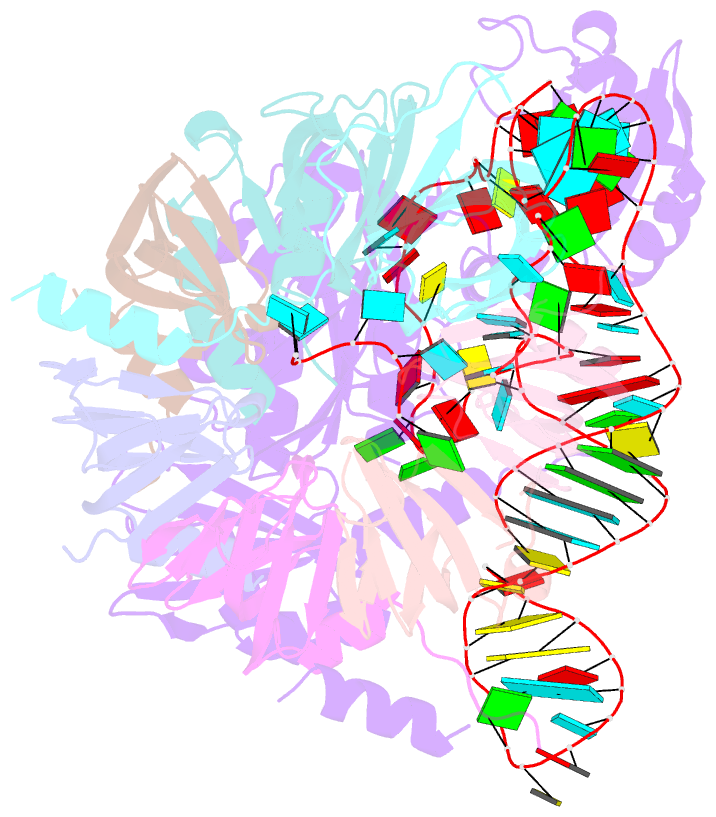
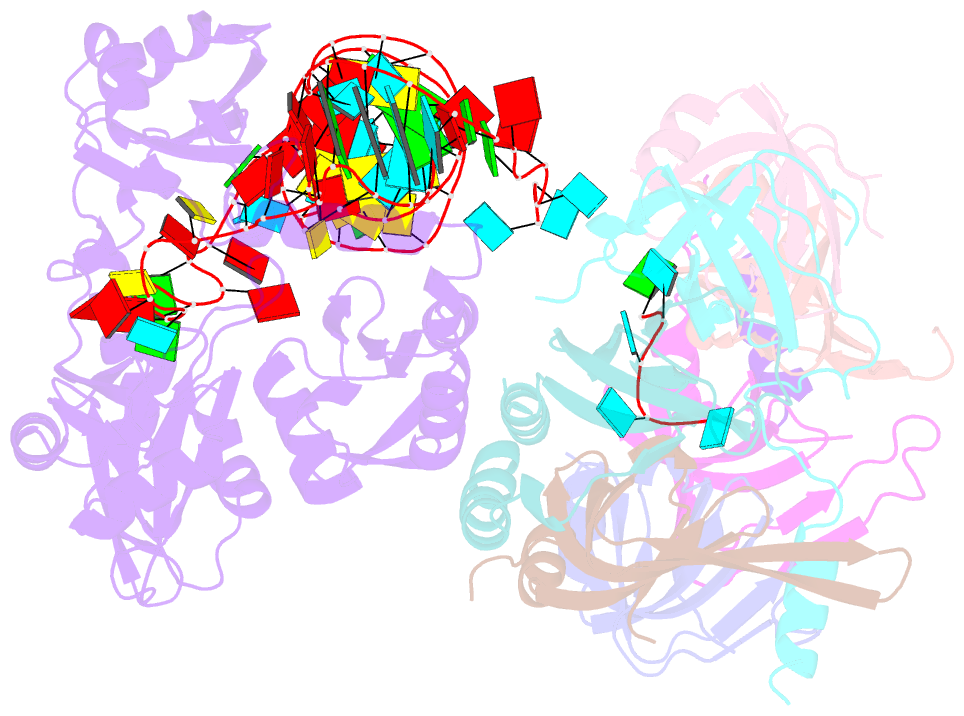
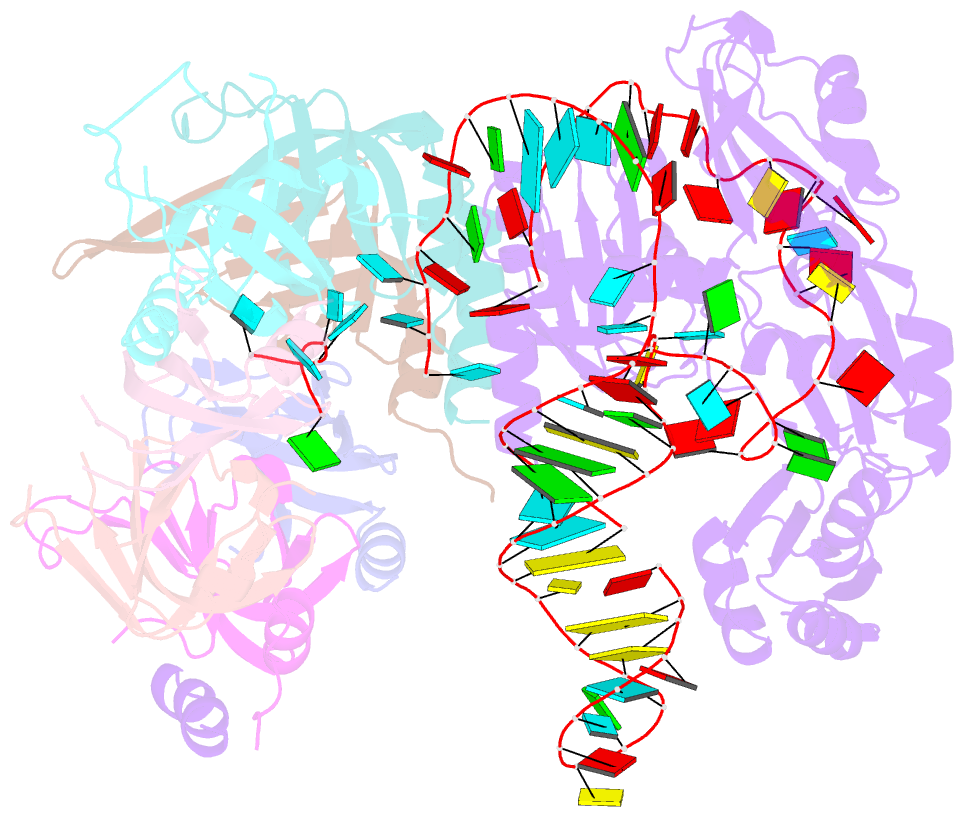
Summary information and primary citation
- PDB-id
- 5vsu; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- splicing
- Method
- X-ray (3.1 Å)
- Summary
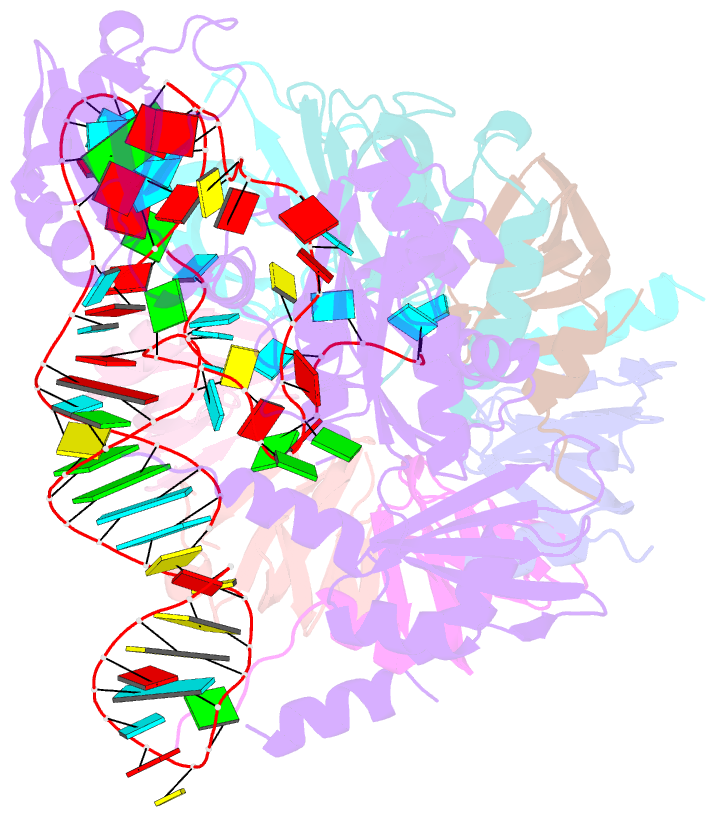
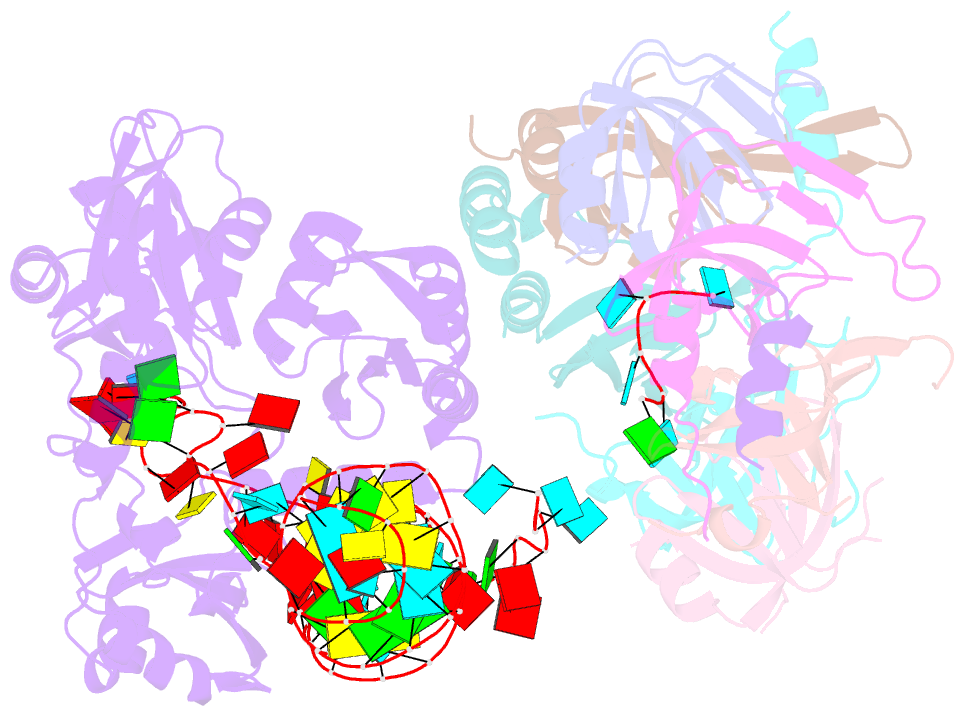
- Structure of yeast u6 snrnp with 2'-phosphate terminated u6 RNA
- Reference
- Montemayor EJ, Didychuk AL, Yake AD, Sidhu GK, Brow DA, Butcher SE (2018): "Architecture of the U6 snRNP reveals specific recognition of 3'-end processed U6 snRNA." Nat Commun, 9, 1749. doi: 10.1038/s41467-018-04145-4.
- Abstract
- The spliceosome removes introns from precursor messenger RNA (pre-mRNA) to produce mature mRNA. Prior to catalysis, spliceosomes are assembled de novo onto pre-mRNA substrates. During this assembly process, U6 small nuclear RNA (snRNA) undergoes extensive structural remodeling. The early stages of this remodeling process are chaperoned by U6 snRNP proteins Prp24 and the Lsm2-8 heteroheptameric ring. We now report a structure of the U6 snRNP from Saccharomyces cerevisiae. The structure reveals protein-protein contacts that position Lsm2-8 in close proximity to the chaperone "active site" of Prp24. The structure also shows how the Lsm2-8 ring specifically recognizes U6 snRNA that has been post-transcriptionally modified at its 3' end, thereby elucidating the mechanism by which U6 snRNPs selectively recruit 3' end-processed U6 snRNA into spliceosomes. Additionally, the structure reveals unanticipated homology between the C-terminal regions of Lsm8 and the cytoplasmic Lsm1 protein involved in mRNA decay.