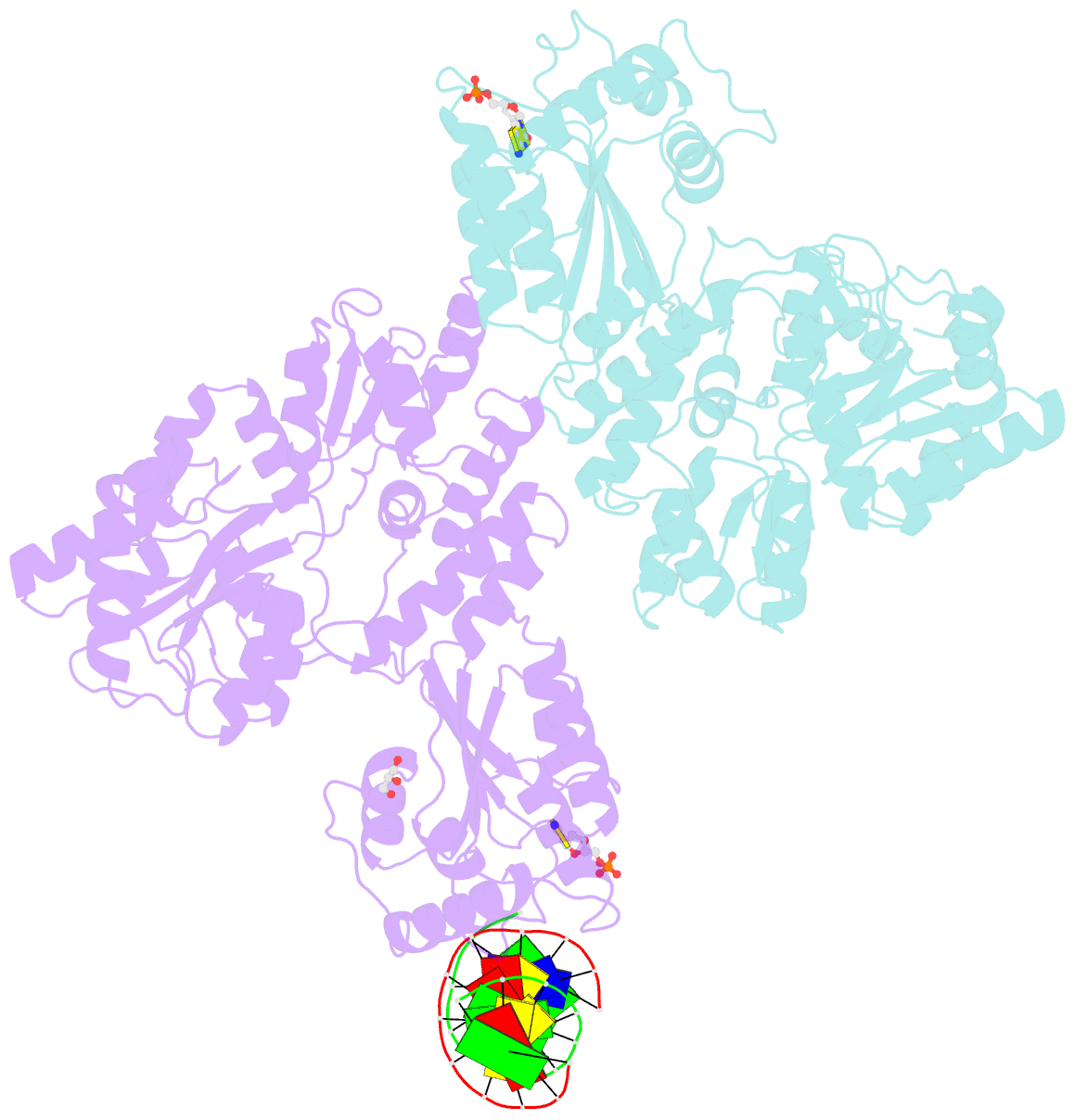
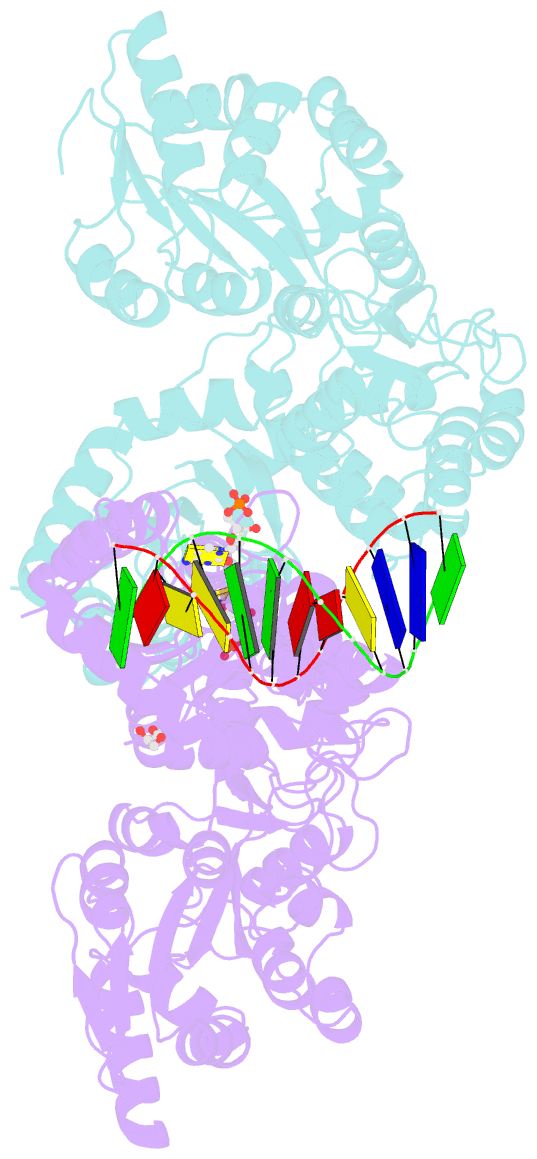
Summary information and primary citation
- PDB-id
- 5w0u; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (2.9 Å)
- Summary
- Crystal structure of mbp fused activation-induced cytidine deaminase (aid) in complex with dcmp
- Reference
- Qiao Q, Wang L, Meng FL, Hwang JK, Alt FW, Wu H (2017): "AID Recognizes Structured DNA for Class Switch Recombination." Mol. Cell, 67, 361-373.e4. doi: 10.1016/j.molcel.2017.06.034.
- Abstract
- Activation-induced cytidine deaminase (AID) initiates both class switch recombination (CSR) and somatic hypermutation (SHM) in antibody diversification. Mechanisms of AID targeting and catalysis remain elusive despite its critical immunological roles and off-target effects in tumorigenesis. Here, we produced active human AID and revealed its preferred recognition and deamination of structured substrates. G-quadruplex (G4)-containing substrates mimicking the mammalian immunoglobulin switch regions are particularly good AID substrates in vitro. By solving crystal structures of maltose binding protein (MBP)-fused AID alone and in complex with deoxycytidine monophosphate, we surprisingly identify a bifurcated substrate-binding surface that explains structured substrate recognition by capturing two adjacent single-stranded overhangs simultaneously. Moreover, G4 substrates induce cooperative AID oligomerization. Structure-based mutations that disrupt bifurcated substrate recognition or oligomerization both compromise CSR in splenic B cells. Collectively, our data implicate intrinsic preference of AID for structured substrates and uncover the importance of G4 recognition and oligomerization of AID in CSR.