Summary information and primary citation
- PDB-id
- 5wea; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-RNA
- Method
- X-ray (3.12 Å)
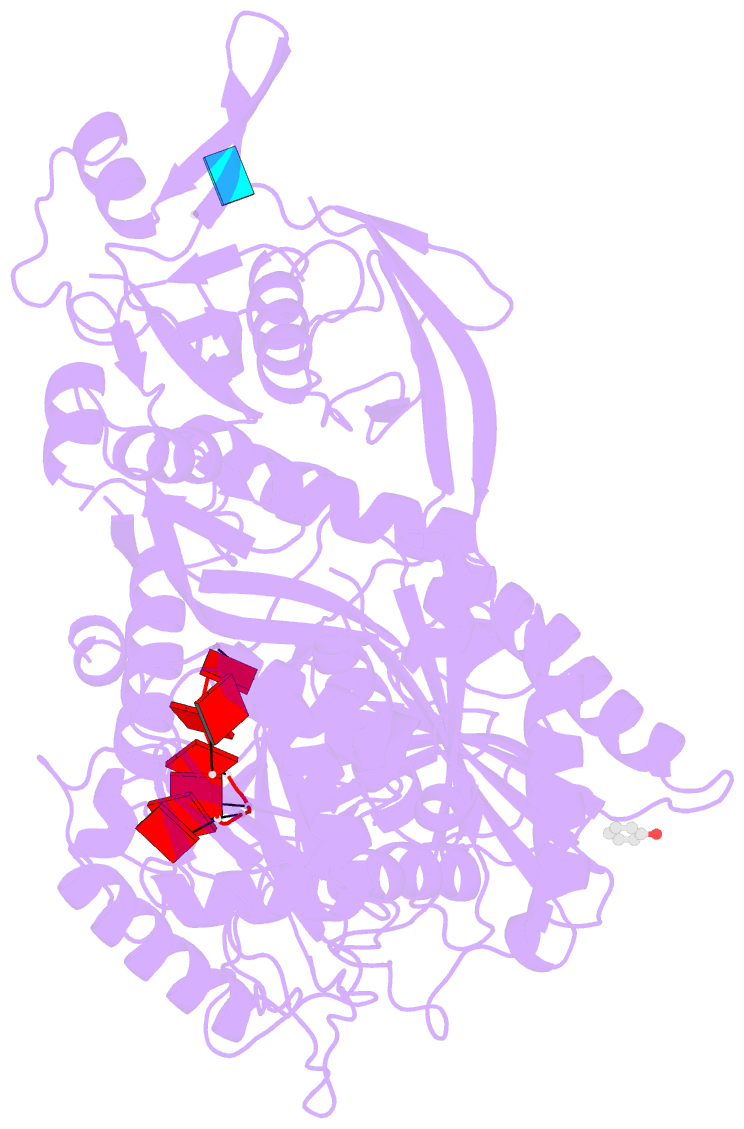
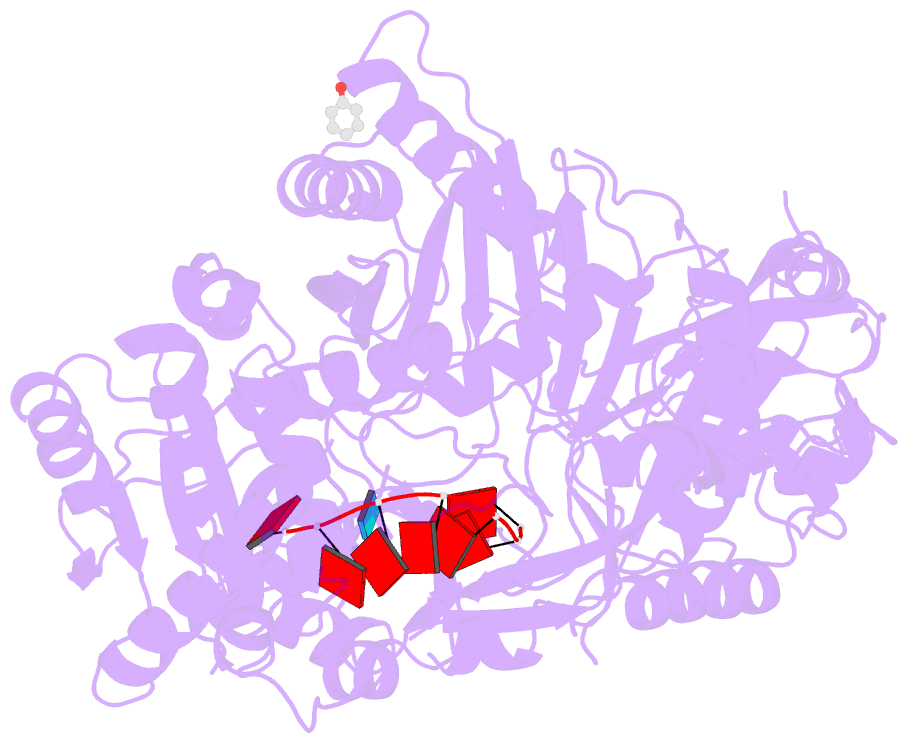
- Summary
- Human argonaute2 helix-7 mutant
- Reference
- Klum SM, Chandradoss SD, Schirle NT, Joo C, MacRae IJ (2018): "Helix-7 in Argonaute2 shapes the microRNA seed region for rapid target recognition." EMBO J., 37, 75-88. doi: 10.15252/embj.201796474.
- Abstract
- Argonaute proteins use microRNAs (miRNAs) to identify mRNAs targeted for post-transcriptional repression. Biochemical assays have demonstrated that Argonaute functions by modulating the binding properties of its miRNA guide so that pairing to the seed region is exquisitely fast and accurate. However, the mechanisms used by Argonaute to reshape the binding properties of its small RNA guide remain poorly understood. Here, we identify a structural element, α-helix-7, in human Argonaute2 (Ago2) that is required for speed and fidelity in binding target RNAs. Biochemical, structural, and single-molecule data indicate that helix-7 acts as a molecular wedge that pivots to enforce rapid making and breaking of miRNA:target base pairs in the 3' half of the seed region. These activities allow Ago2 to rapidly dismiss off-targets and dynamically search for seed-matched sites at a rate approaching the limit of diffusion.