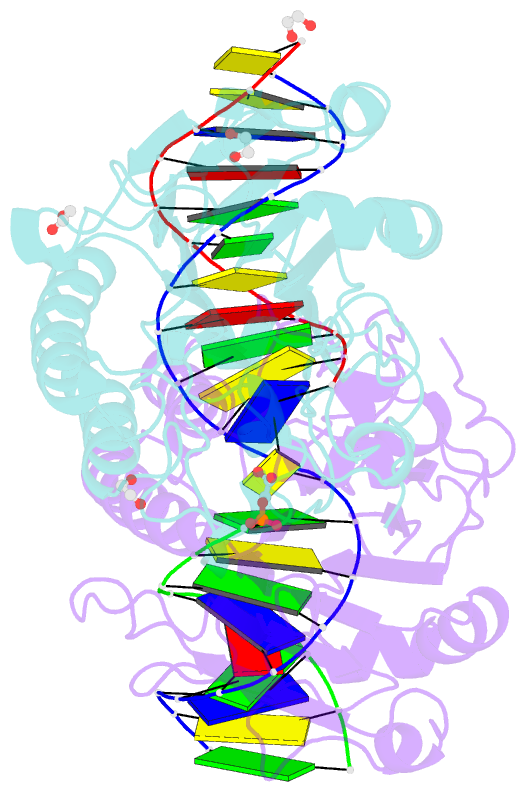
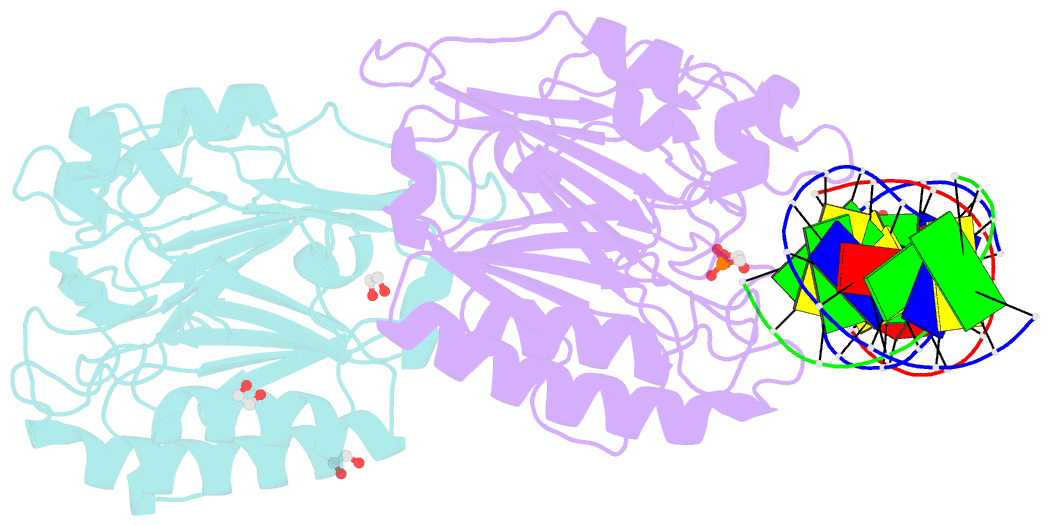
Summary information and primary citation
- PDB-id
- 5wn2; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase, lyase-DNA
- Method
- X-ray (2.288 Å)
- Summary
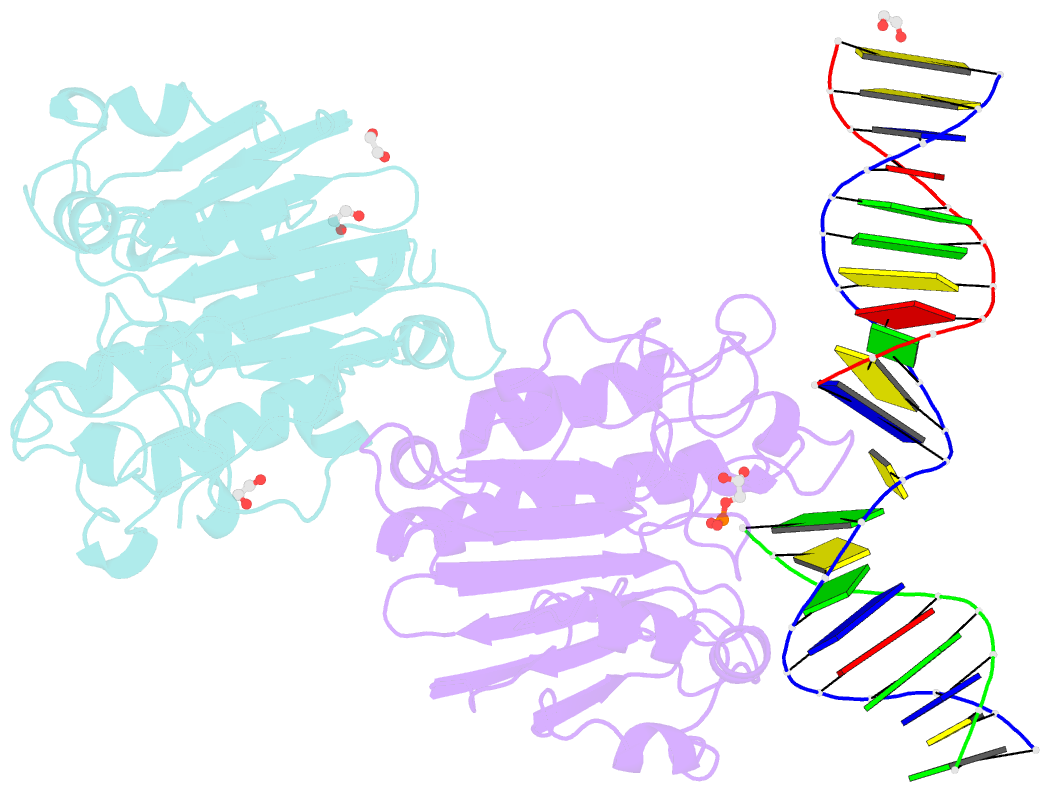
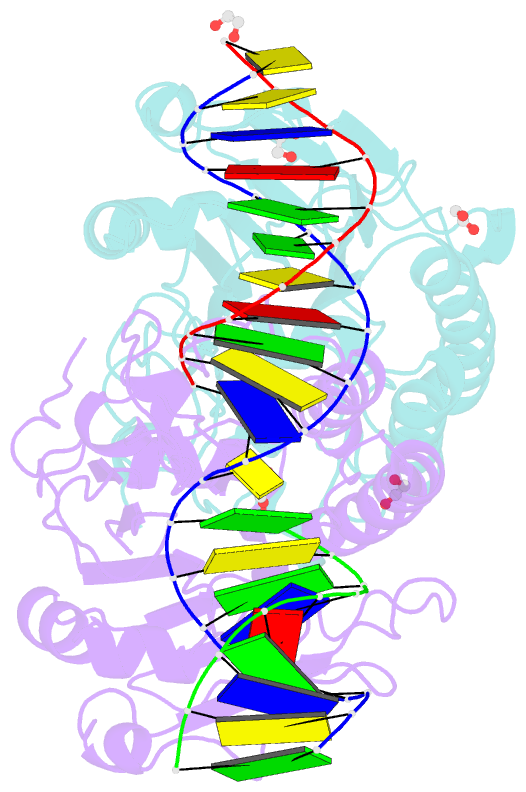
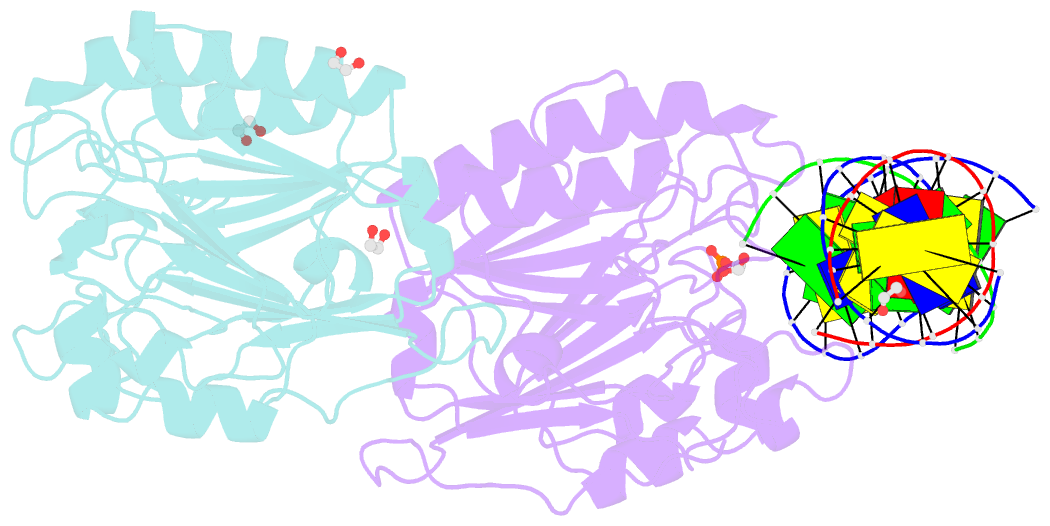
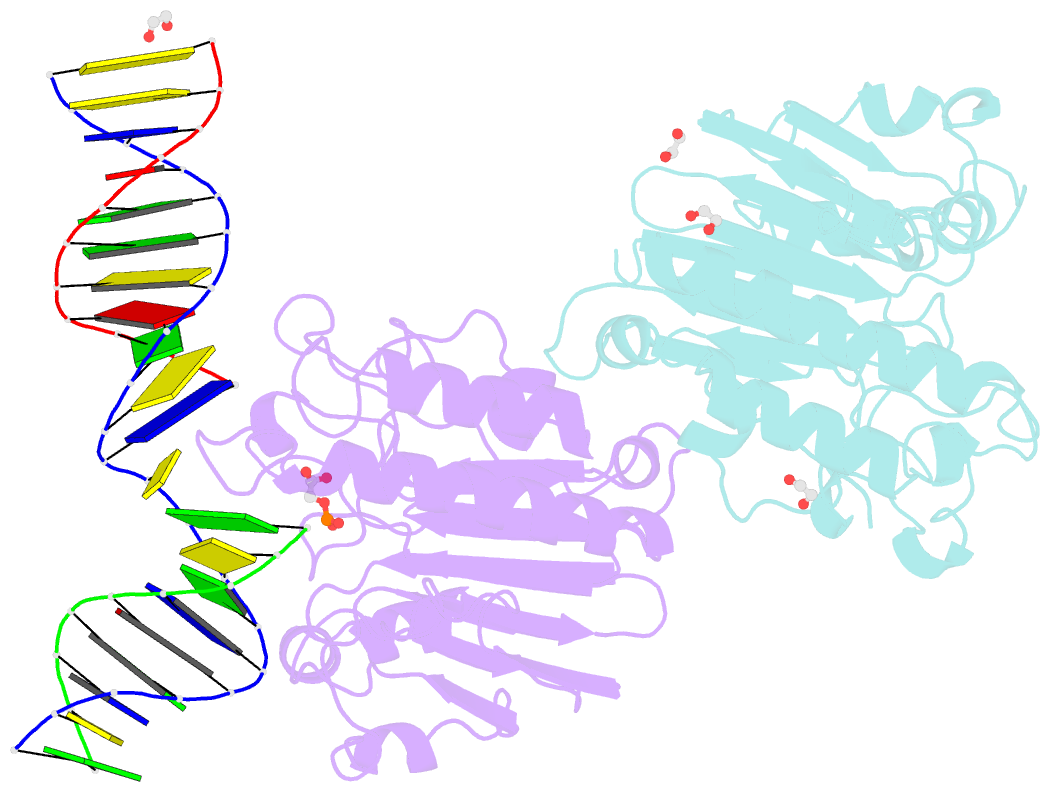
- Ape1 exonuclease substrate complex with phosphoglycolate
- Reference
- Whitaker AM, Flynn TS, Freudenthal BD (2018): "Molecular snapshots of APE1 proofreading mismatches and removing DNA damage." Nat Commun, 9, 399. doi: 10.1038/s41467-017-02175-y.
- Abstract
- Human apurinic/apyrimidinic (AP) endonuclease 1 (APE1) is an essential DNA repair enzyme which uses a single active site to process DNA damage via two distinct activities: (1) AP-endonuclease and (2) 3' to 5' exonuclease. The AP-endonuclease activity cleaves at AP-sites, while the exonuclease activity excises bulkier 3' mismatches and DNA damage to generate clean DNA ends suitable for downstream repair. Molecular details of the exonuclease reaction and how one active site can accommodate various toxic DNA repair intermediates remains elusive despite being biologically important. Here, we report multiple high-resolution APE1-DNA structural snapshots revealing how APE1 removes 3' mismatches and DNA damage by placing the 3' group within the intra-helical DNA cavity via a non-base flipping mechanism. This process is facilitated by a DNA nick, instability of a mismatched/damaged base, and bending of the DNA. These results illustrate how APE1 cleanses DNA dirty-ends to generate suitable substrates for downstream repair enzymes.