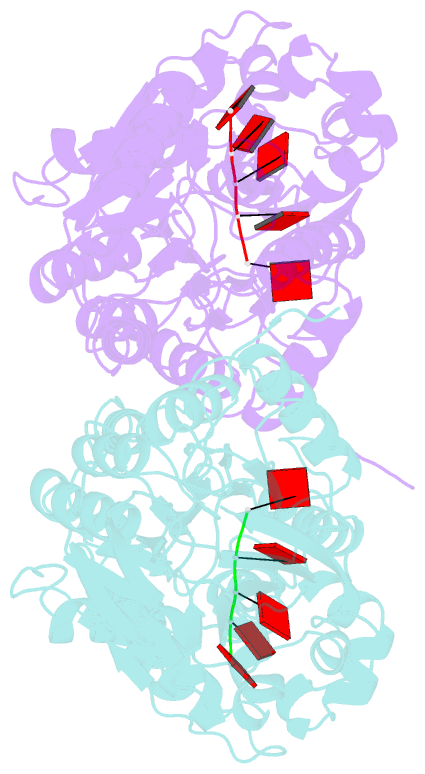
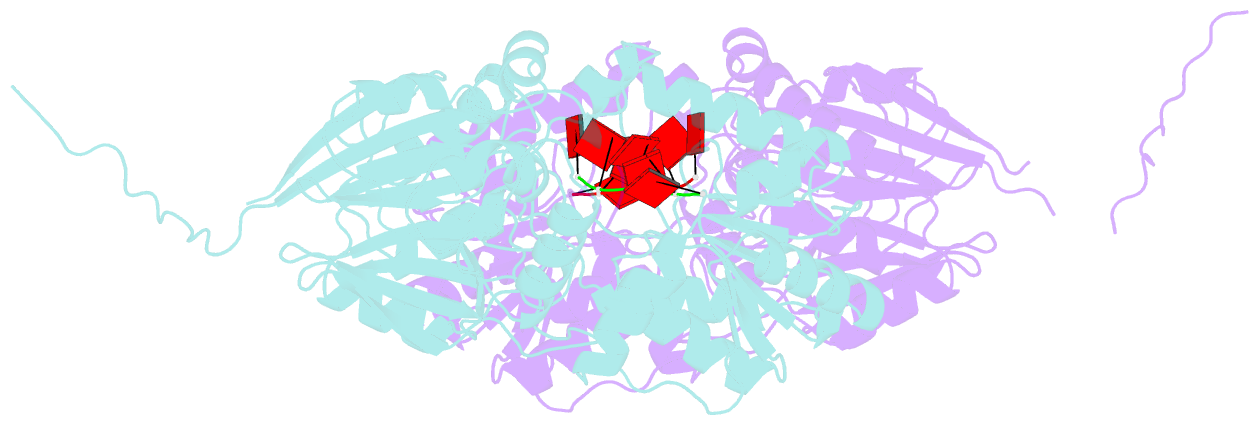
Summary information and primary citation
- PDB-id
- 5ws2; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase
- Method
- X-ray (2.398 Å)
- Summary
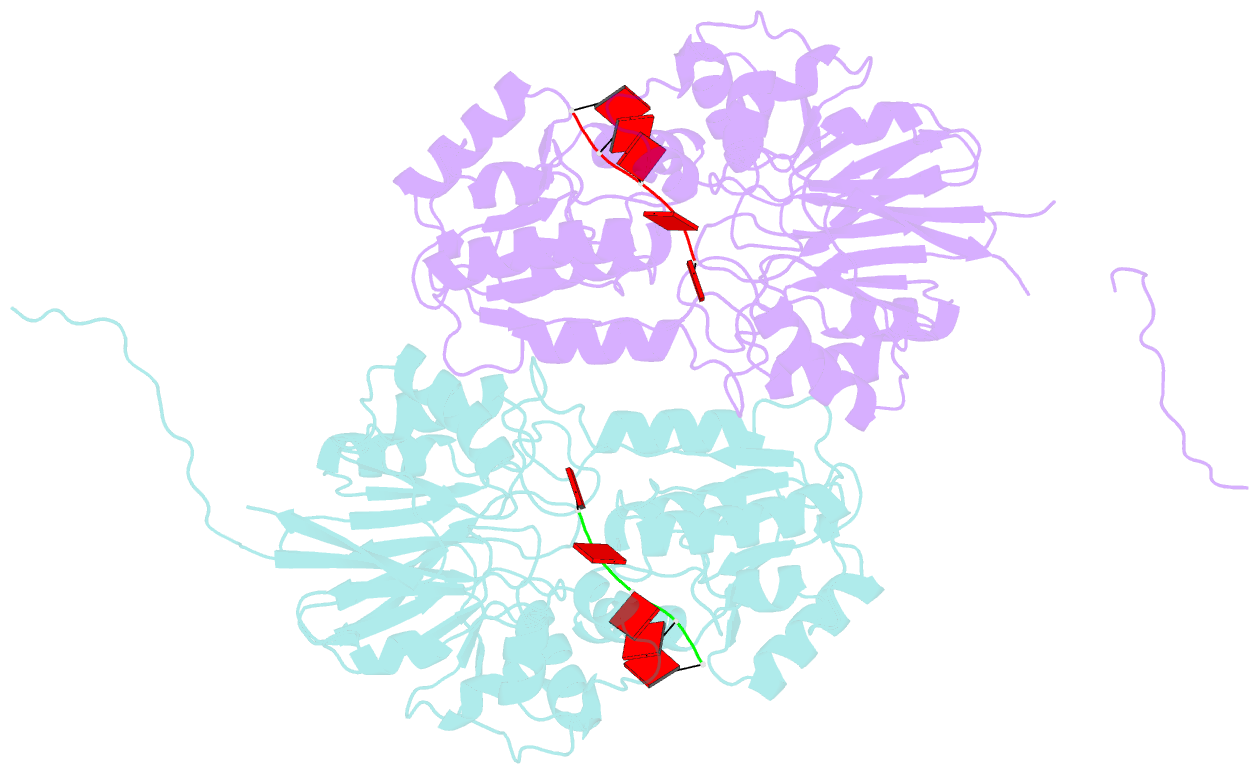
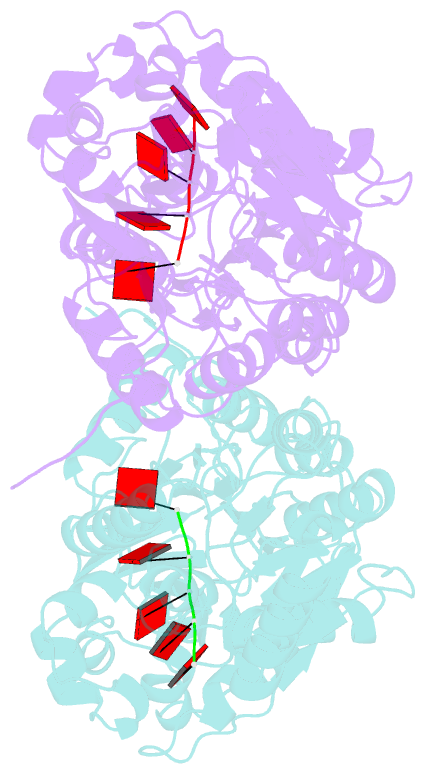
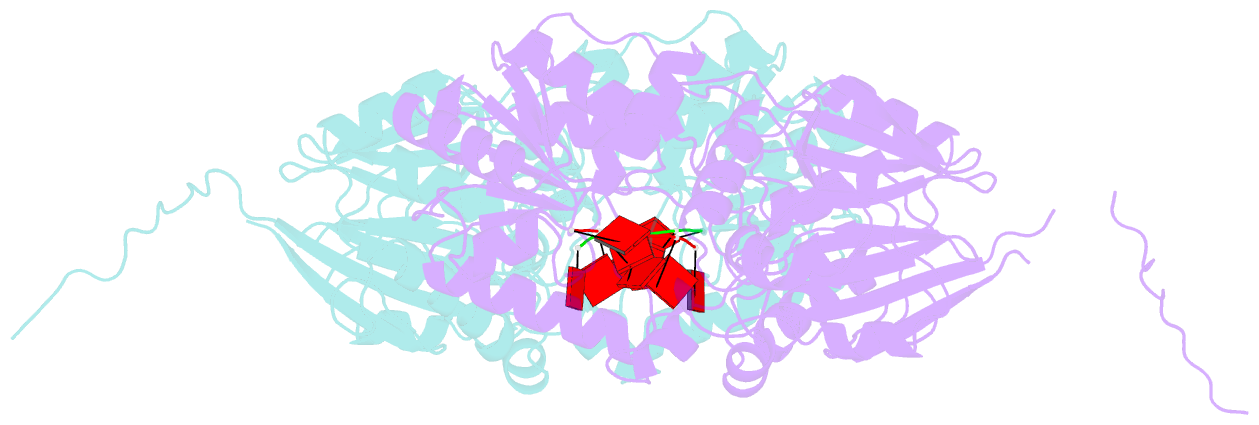
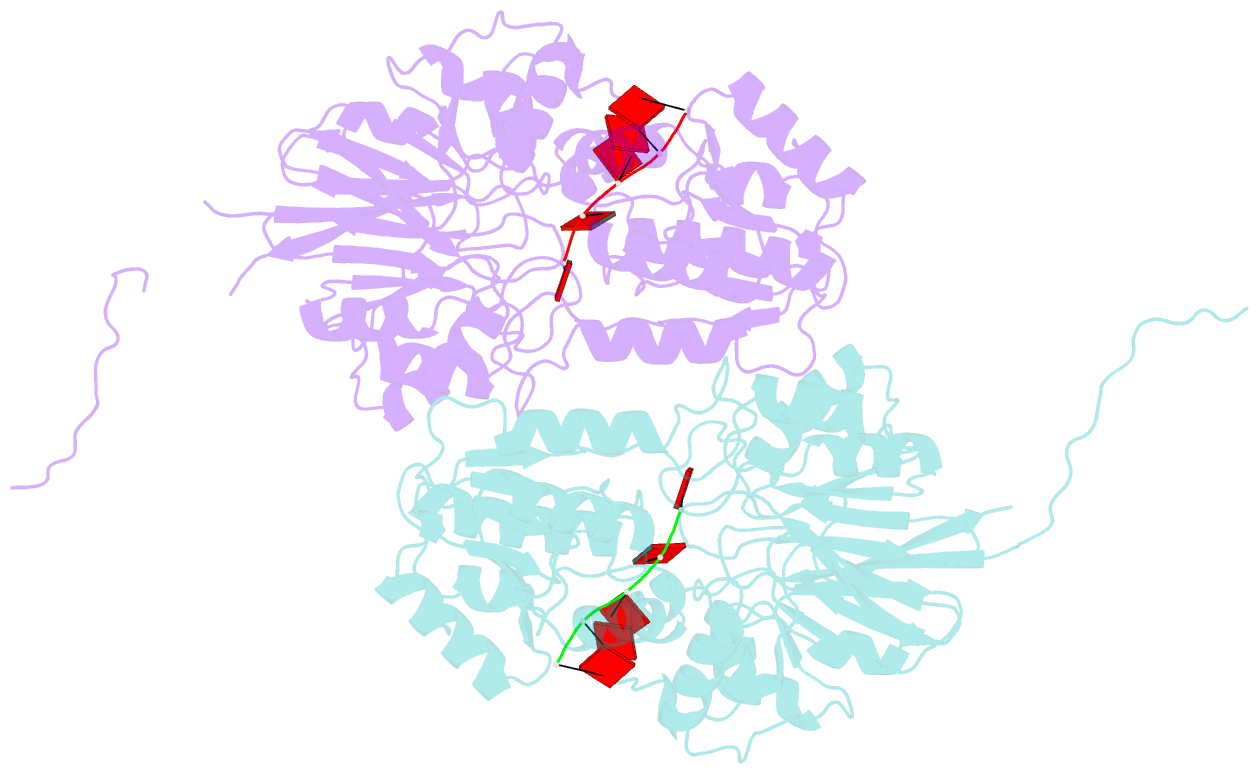
- Crystal structure of mpy-rnase j (mutant s247a), an archaeal rnase j from methanolobus psychrophilus r15, complex with RNA
- Reference
- Zheng X, Feng N, Li D, Dong X, Li J (2017): "New molecular insights into an archaeal RNase J reveal a conserved processive exoribonucleolysis mechanism of the RNase J family." Mol. Microbiol., 106, 351-366. doi: 10.1111/mmi.13769.
- Abstract
- RNase J, a prokaryotic 5'-3' exo/endoribonuclease, contributes to mRNA decay, rRNA maturation and post-transcriptional regulation. Yet the processive-exoribonucleolysis mechanism remains obscure. Here, we solved the first RNA-free and RNA-bound structures of an archaeal RNase J, and through intensive biochemical studies provided detailed mechanistic insights into the catalysis and processivity. Distinct dimerization/tetramerization patterns were observed for archaeal and bacterial RNase Js, and unique archaeal Loops I and II were found involved in RNA interaction. A hydrogen-bond-network was identified for the first time that assists catalysis by facilitating efficient proton transfer in the catalytic center. A conserved 5'-monophosphate-binding pocket that coordinates the RNA 5'-end ensures the 5'-monophosphate preferential exoribonucleolysis. To achieve exoribonucleolytic processivity, the 5'-monophosphate-binding pocket and nucleotide +4 binding site anchor RNA within the catalytic track; the 5'-capping residue Leu37 of the sandwich pocket coupled with the 5'-monophosphate-binding pocket are dedicated to translocating and controlling the RNA orientation for each exoribonucleolytic cycle. The processive-exoribonucleolysis mechanism was verified as conserved in bacterial RNase J and also exposes striking parallels with the non-homologous eukaryotic 5'-3' exoribonuclease, Xrn1. The findings in this work shed light on not only the molecular mechanism of the RNase J family, but also the evolutionary convergence of divergent exoribonucleases.