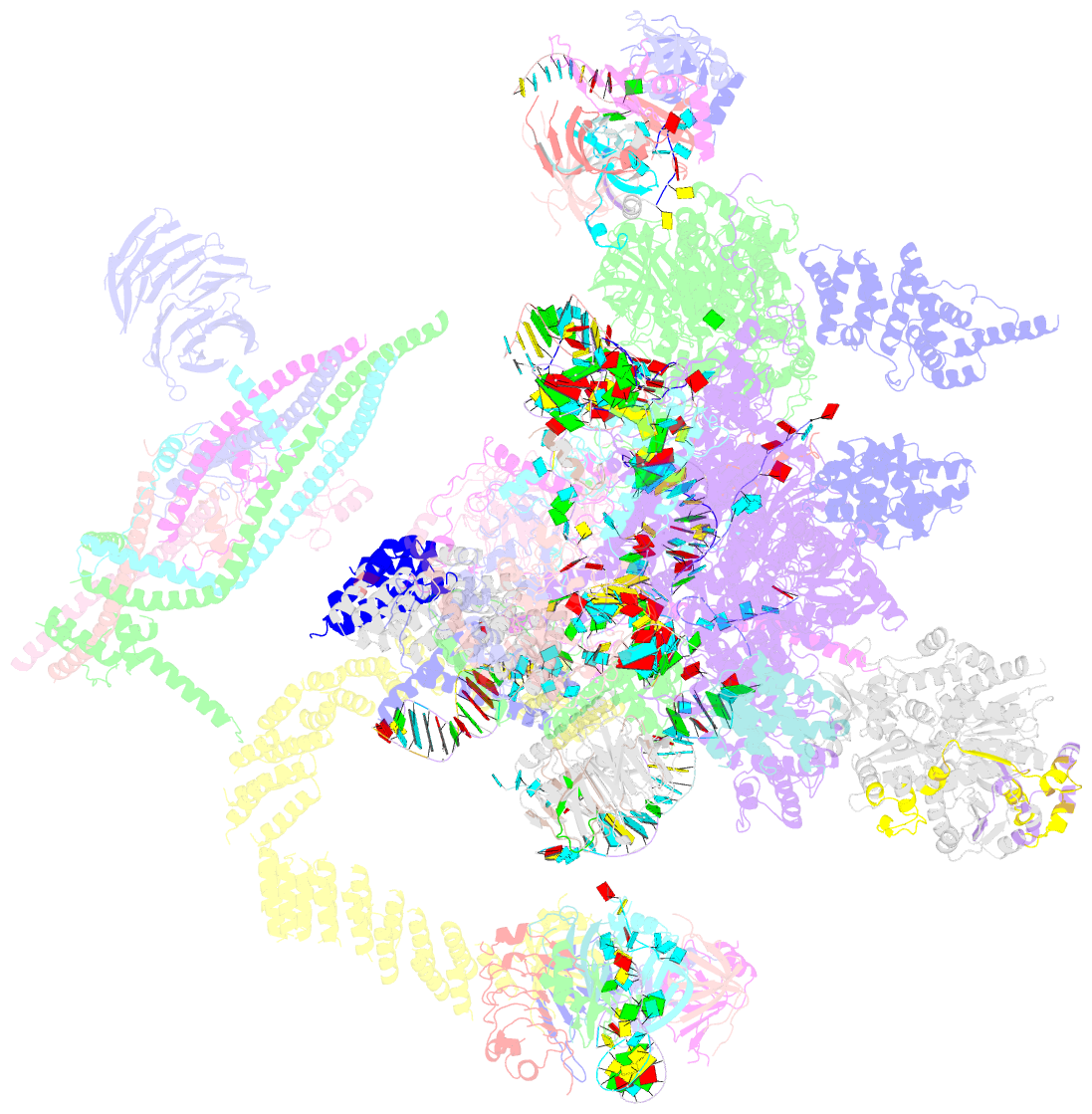
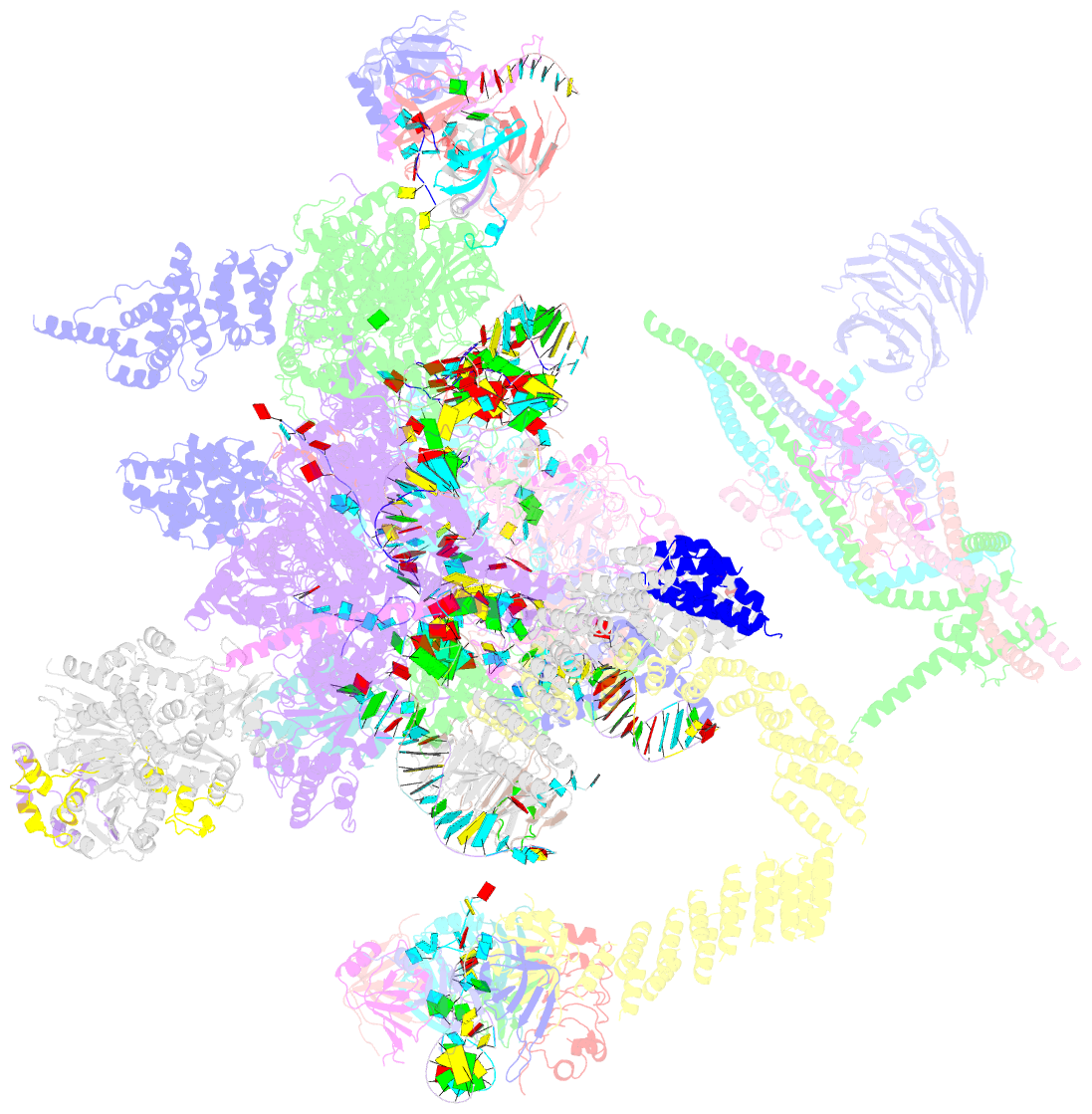
Summary information and primary citation
- PDB-id
- 5wsg; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- cryo-EM (4.0 Å)
- Summary
- cryo-EM structure of the catalytic step ii spliceosome (c* complex) at 4.0 angstrom resolution
- Reference
- Yan C, Wan R, Bai R, Huang G, Shi Y (2017): "Structure of a yeast step II catalytically activated spliceosome." Science, 355, 149-155. doi: 10.1126/science.aak9979.
- Abstract
- Each cycle of precursor messenger RNA (pre-mRNA) splicing comprises two sequential reactions, first freeing the 5' exon and generating an intron lariat-3' exon and then ligating the two exons and releasing the intron lariat. The second reaction is executed by the step II catalytically activated spliceosome (known as the C* complex). Here, we present the cryo-electron microscopy structure of a C* complex from Saccharomyces cerevisiae at an average resolution of 4.0 angstroms. Compared with the preceding spliceosomal complex (C complex), the lariat junction has been translocated by 15 to 20 angstroms to vacate space for the incoming 3'-exon sequences. The step I splicing factors Cwc25 and Yju2 have been dissociated from the active site. Two catalytic motifs from Prp8 (the 1585 loop and the β finger of the ribonuclease H-like domain), along with the step II splicing factors Prp17 and Prp18 and other surrounding proteins, are poised to assist the second transesterification. These structural features, together with those reported for other spliceosomal complexes, yield a near-complete mechanistic picture on the splicing cycle.