Summary information and primary citation
- PDB-id
- 5wwc; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (1.9 Å)
- Summary
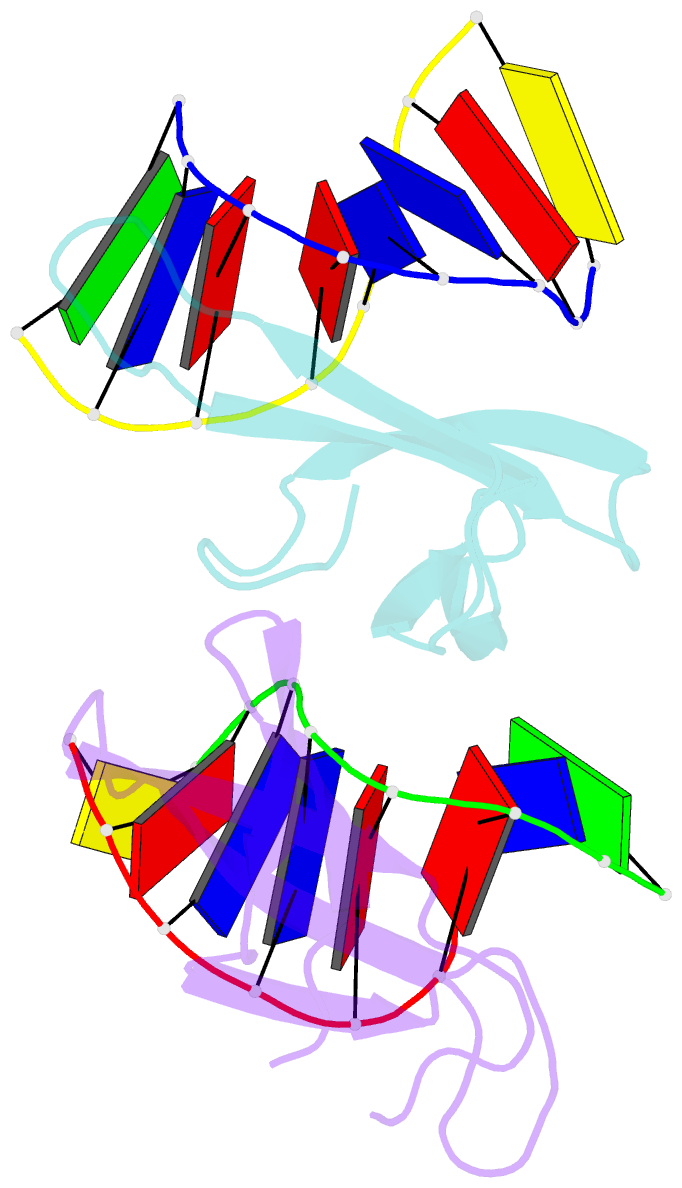
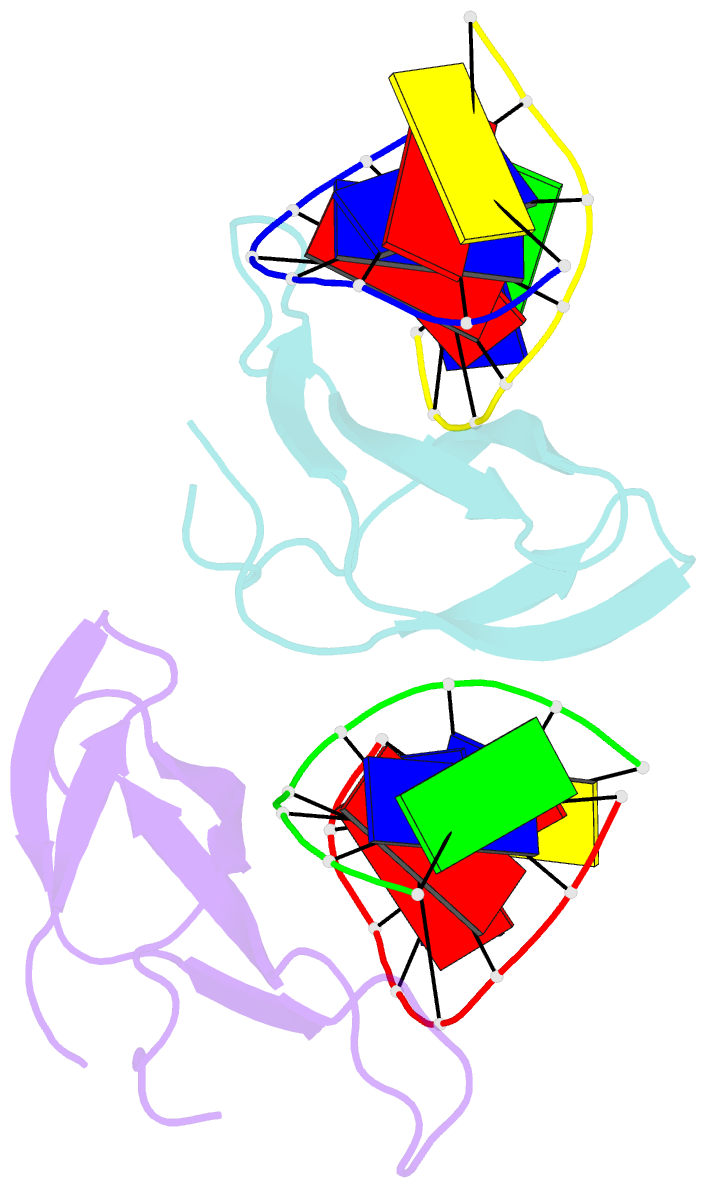
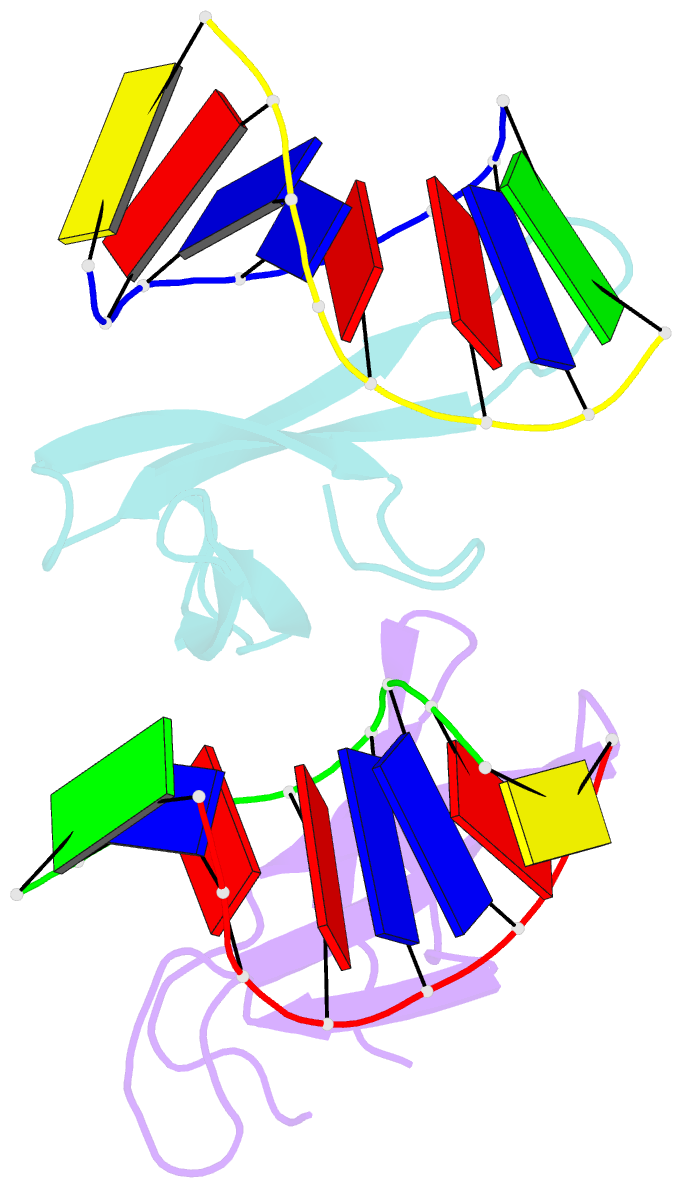
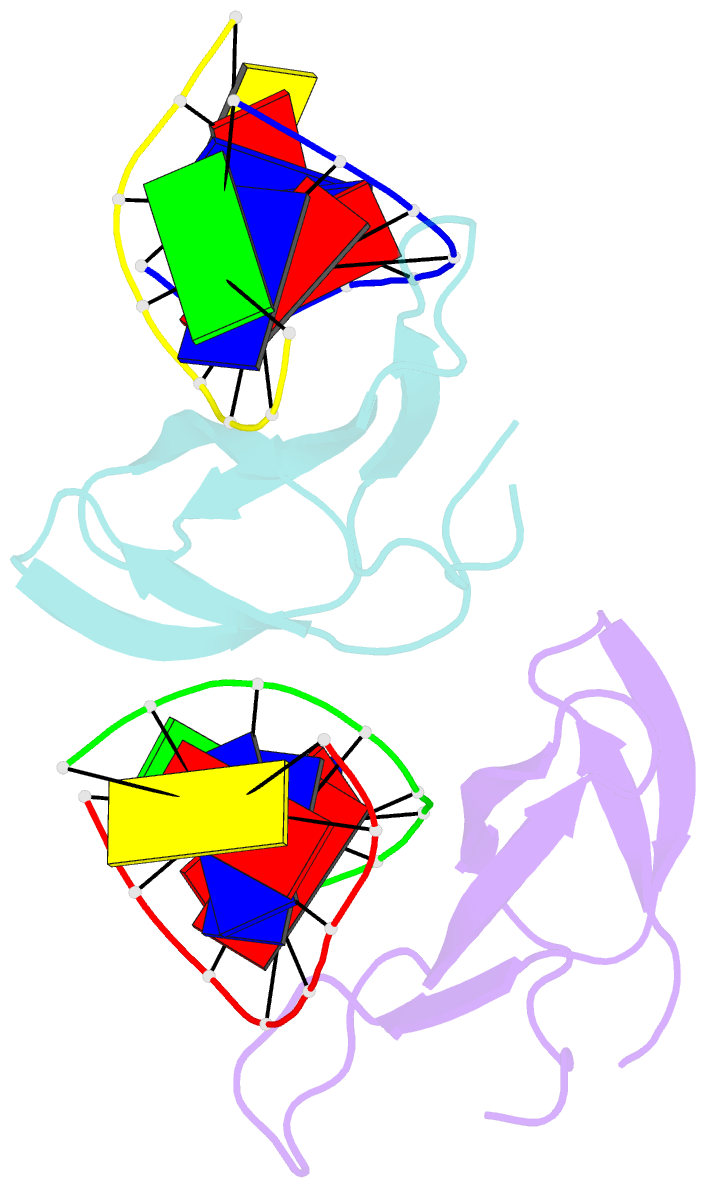
- The crystal structure of cren7 mutant l28m in complex with dsDNA
- Reference
- Zhang Z, Zhao M, Wang L, Chen Y, Dong Y, Gong Y, Huang L (2017): "Roles of Leu28 side chain intercalation in the interaction between Cren7 and DNA." Biochem. J., 474, 1727-1739. doi: 10.1042/BCJ20170036.
- Abstract
- Crenarchaeal chromatin protein Cren7 binds double-stranded DNA in the minor groove, introducing a sharp single-step DNA kink. The side chain of Leu28, a residue conserved among all Cren7 homologs, intercalates into the kinked DNA step. In the present study, we replaced Leu28 with a residue containing a hydrophobic side chain of different sizes (i.e. L28A, L28V, L28I, L28M and L28F). Both the stability of the Cren7-DNA complex and the ability of Cren7 to constrain DNA supercoils correlated well with the size of the intercalated side chain. Structural analysis shows that L28A induces a kink (∼43°), nearly as sharp as that produced by wild-type Cren7 (∼48°), in the bound DNA fragment despite the lack of side chain intercalation. In another duplex DNA fragment, L28F inserts a large hydrophobic side chain deep into the DNA step, but introduces a smaller kink (∼39°) than that formed by the wild-type protein (∼50°). Mutation of Leu28 into methionine yields two protein conformers differing in loop β3-β4 orientation, DNA-binding surface and DNA geometry in the protein-DNA structure. Our results indicate that side chain intercalation is not directly responsible for DNA kinking or bending by Cren7, but plays a critical role in the stabilization of the Cren7-DNA complex. In addition, the flexibility of loop β3-β4 in Cren7, as revealed in the crystal structure of L28M-DNA, may serve a role in the modulation of chromosomal organization and function in the cell.