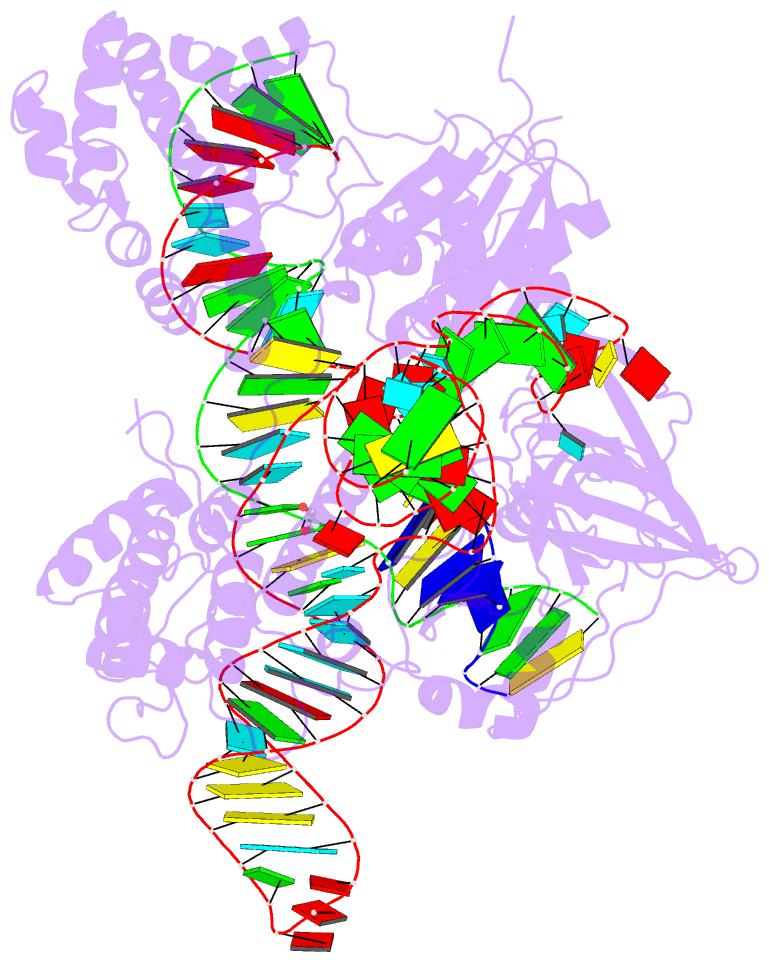
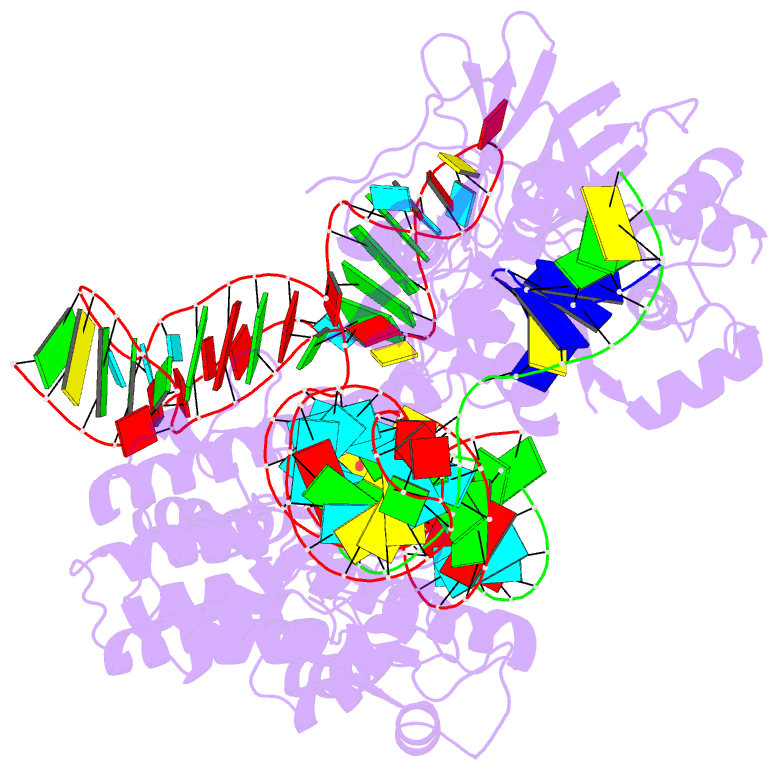
Summary information and primary citation
- PDB-id
- 5x2g; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-RNA-DNA
- Method
- X-ray (2.4 Å)
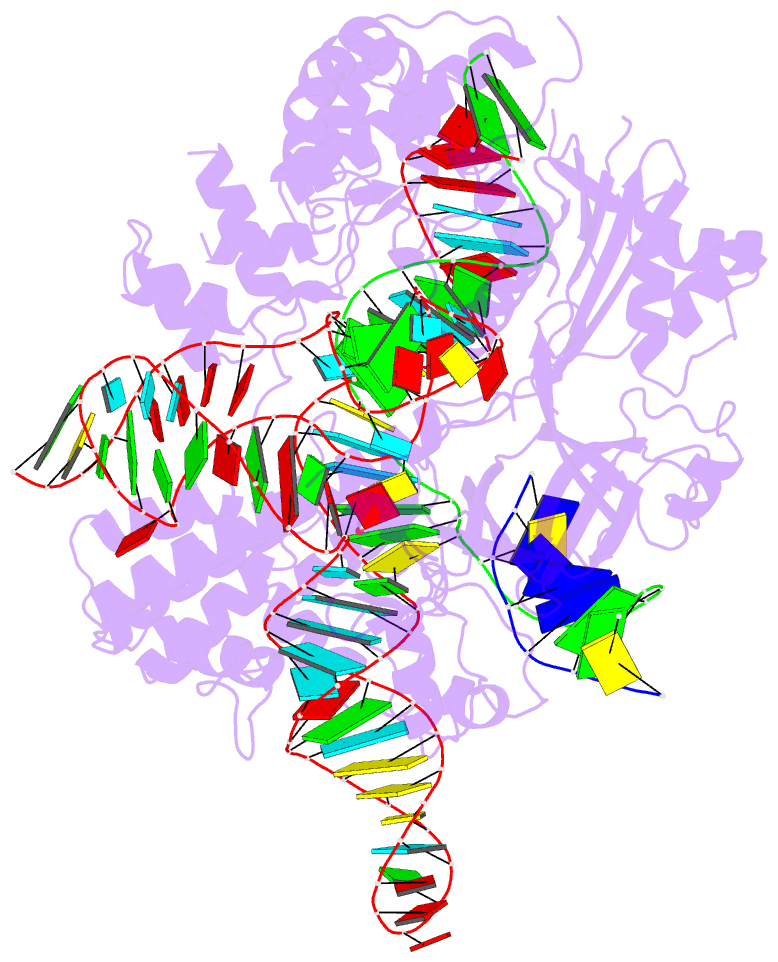
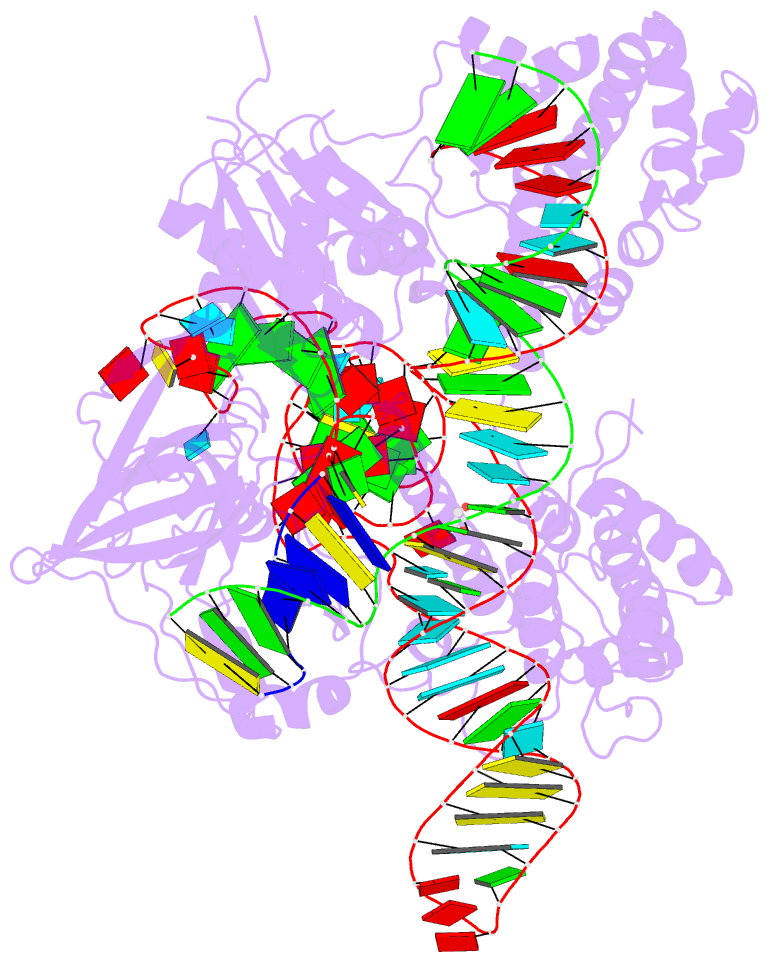
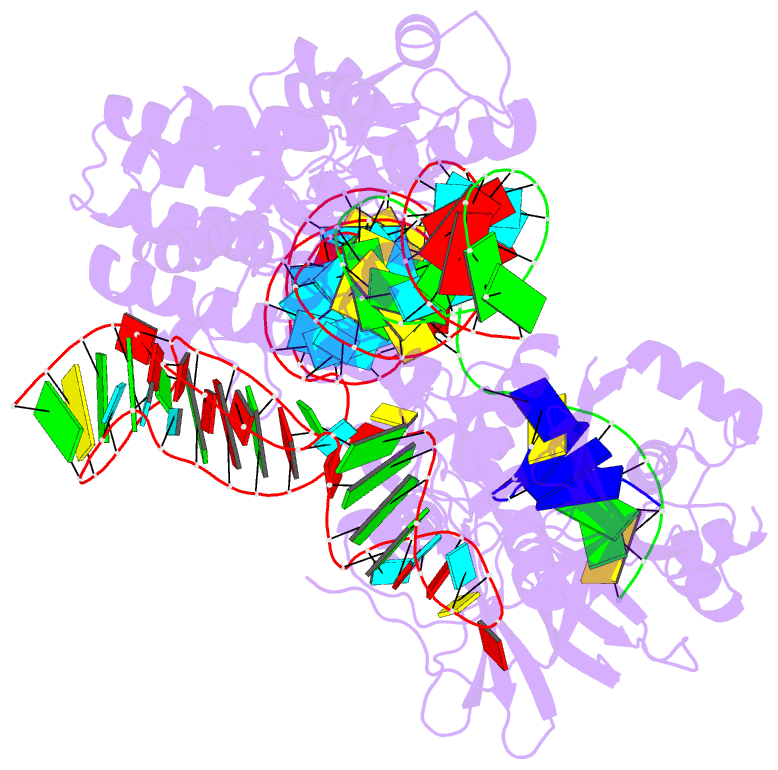
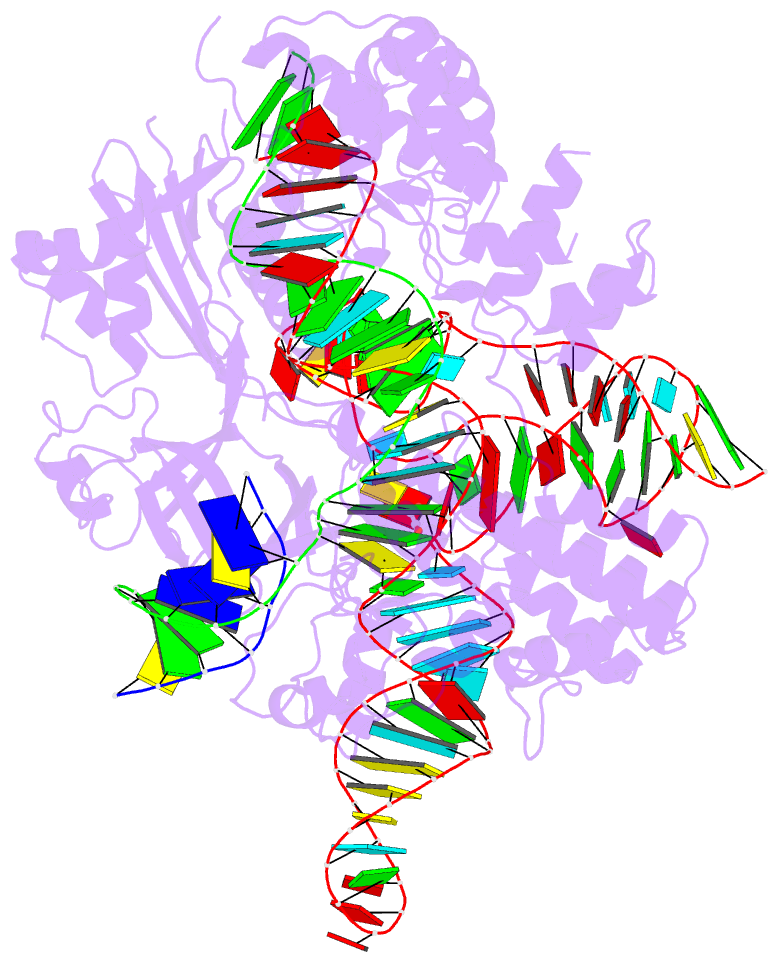
- Summary
- Crystal structure of campylobacter jejuni cas9 in complex with sgrna and target DNA (agaaacc pam)
- Reference
- Yamada M, Watanabe Y, Gootenberg JS, Hirano H, Ran FA, Nakane T, Ishitani R, Zhang F, Nishimasu H, Nureki O (2017): "Crystal Structure of the Minimal Cas9 from Campylobacter jejuni Reveals the Molecular Diversity in the CRISPR-Cas9 Systems." Mol. Cell, 65, 1109-1121.e3. doi: 10.1016/j.molcel.2017.02.007.
- Abstract
- The RNA-guided endonuclease Cas9 generates a double-strand break at DNA target sites complementary to the guide RNA and has been harnessed for the development of a variety of new technologies, such as genome editing. Here, we report the crystal structures of Campylobacter jejuni Cas9 (CjCas9), one of the smallest Cas9 orthologs, in complex with an sgRNA and its target DNA. The structures provided insights into a minimal Cas9 scaffold and revealed the remarkable mechanistic diversity of the CRISPR-Cas9 systems. The CjCas9 guide RNA contains a triple-helix structure, which is distinct from known RNA triple helices, thereby expanding the natural repertoire of RNA triple helices. Furthermore, unlike the other Cas9 orthologs, CjCas9 contacts the nucleotide sequences in both the target and non-target DNA strands and recognizes the 5'-NNNVRYM-3' as the protospacer-adjacent motif. Collectively, these findings improve our mechanistic understanding of the CRISPR-Cas9 systems and may facilitate Cas9 engineering.