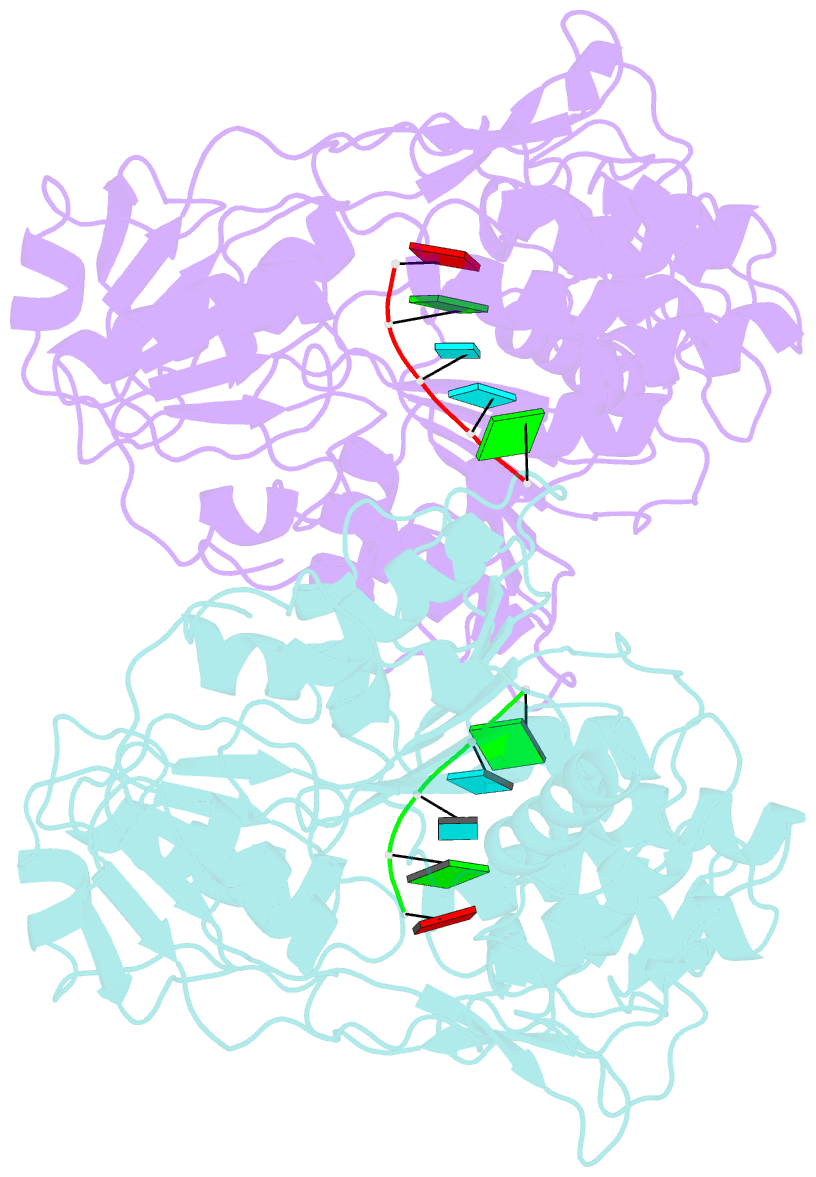
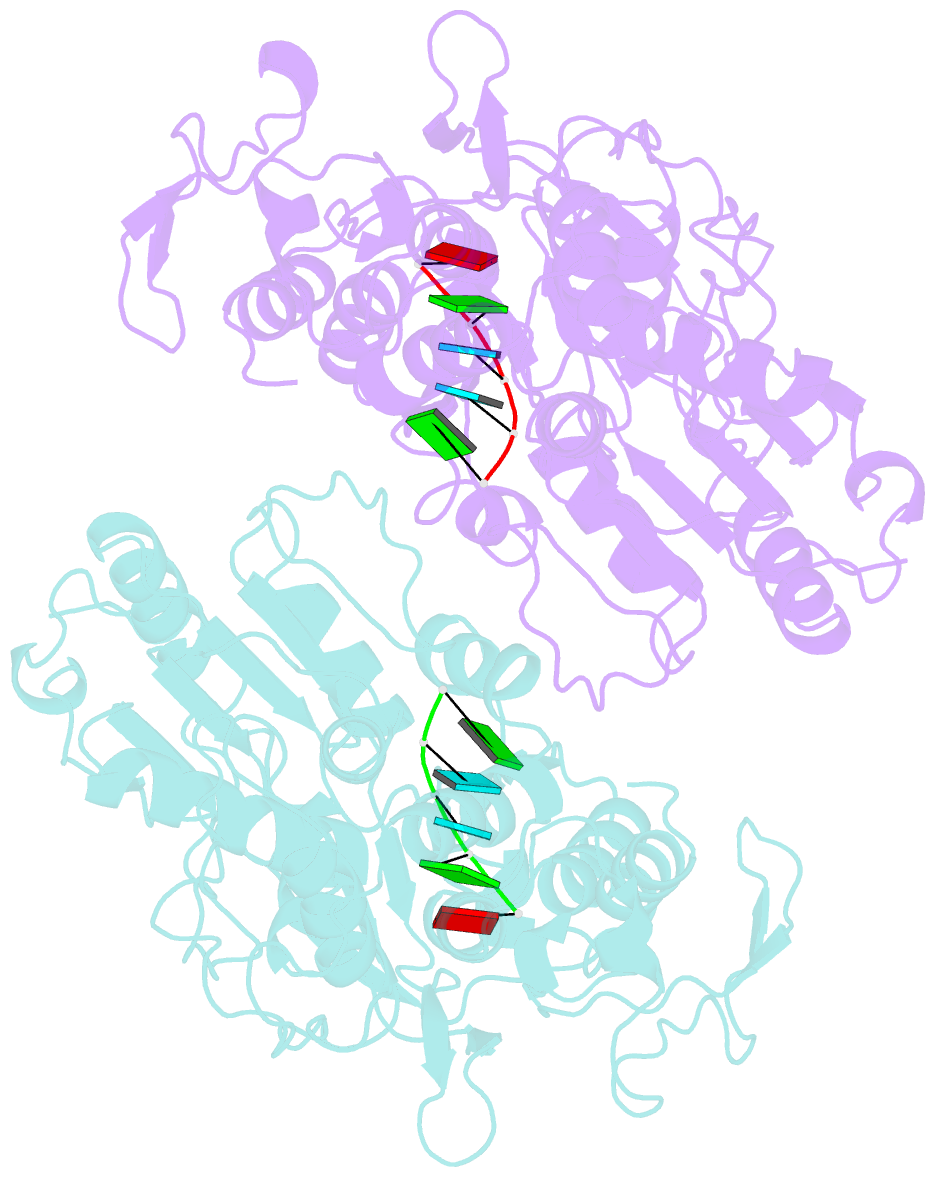
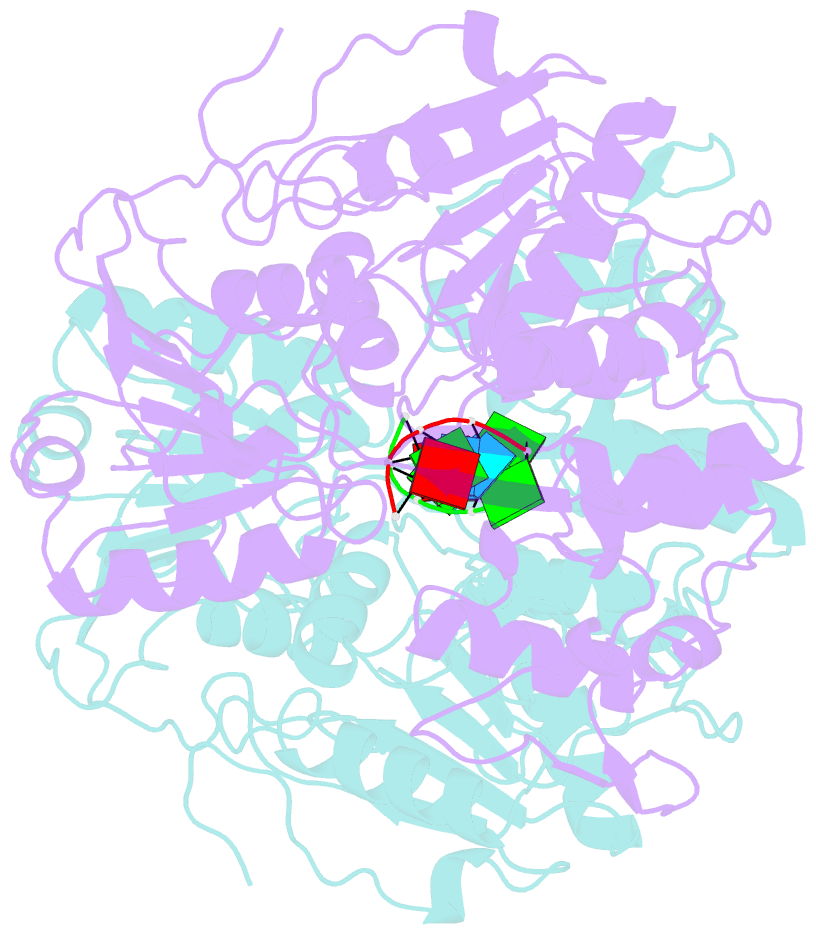
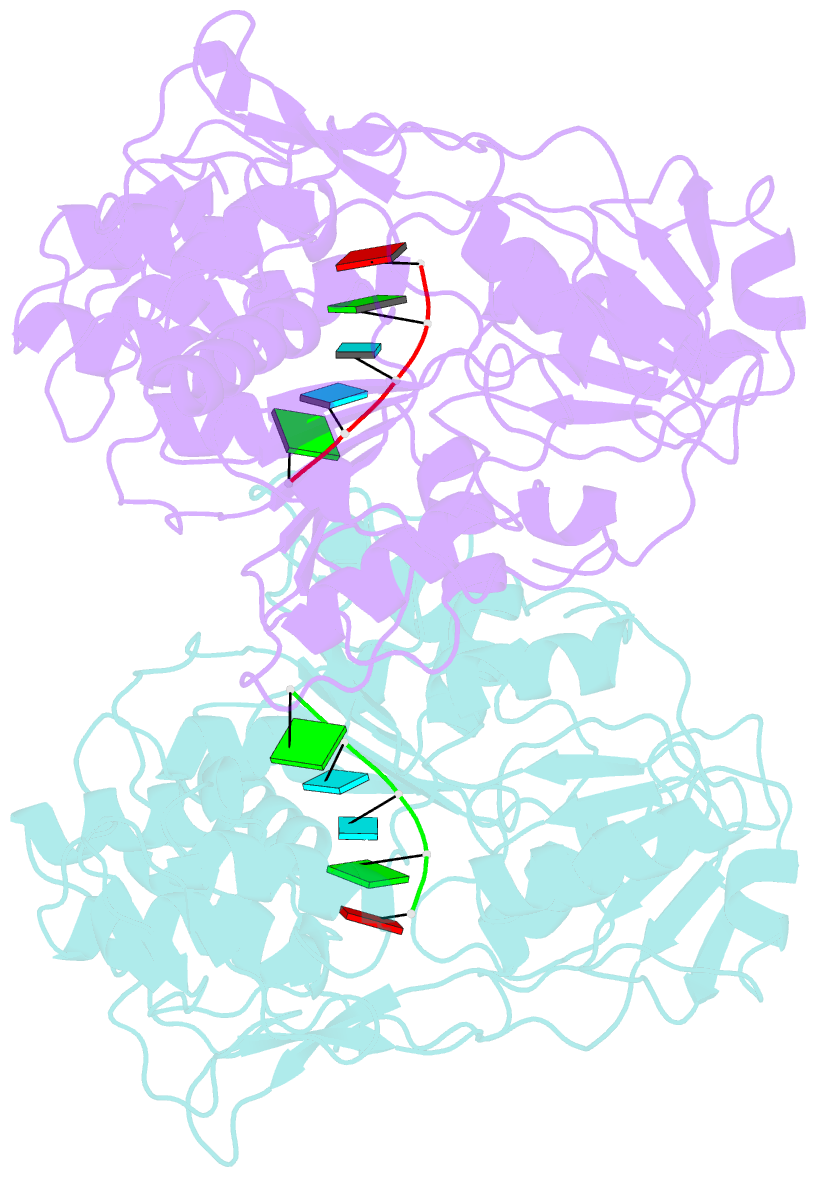
Summary information and primary citation
- PDB-id
- 5xc6; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-RNA
- Method
- X-ray (2.9 Å)
- Summary
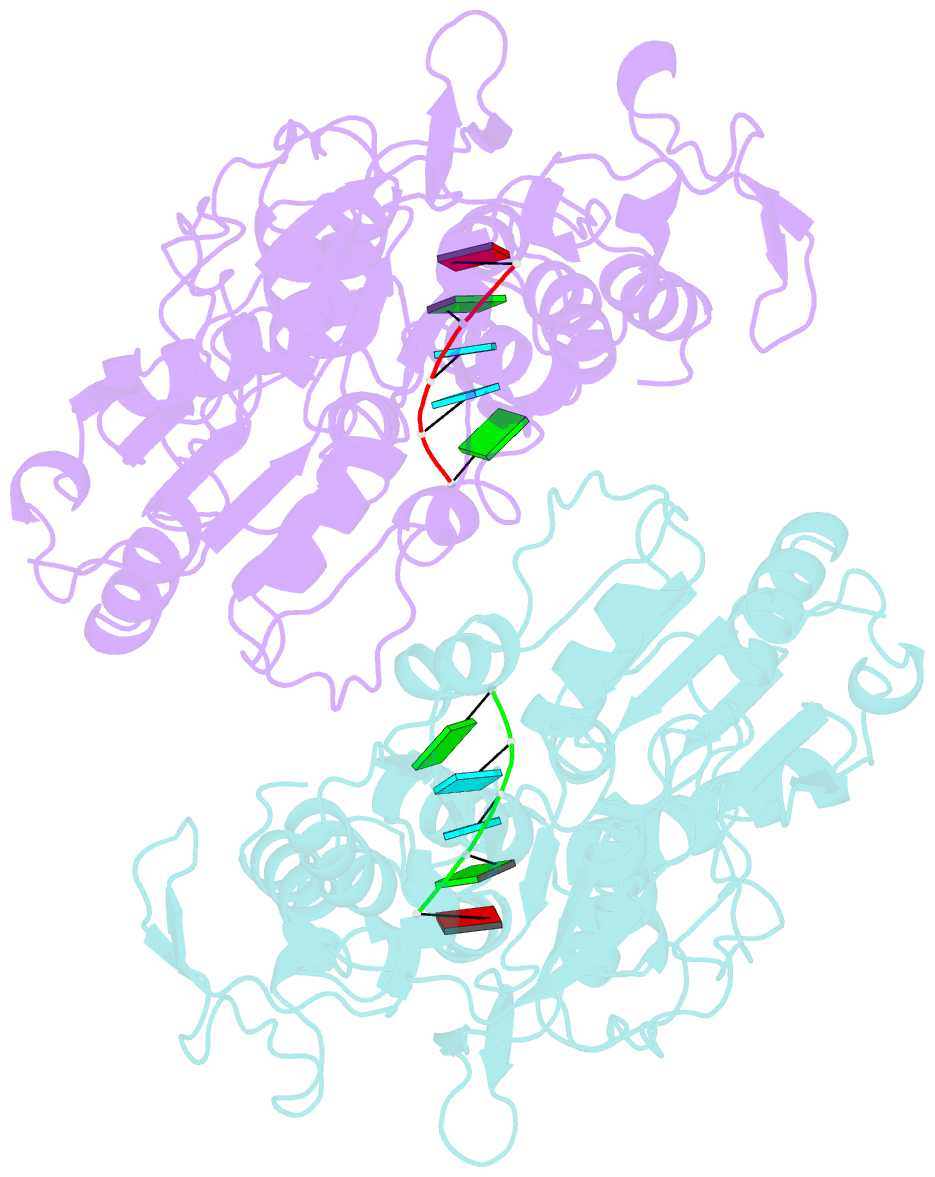
- Dengue virus 4 ns3 helicase in complex with ssrna sla12
- Reference
- Swarbrick CMD, Basavannacharya C, Chan KWK, Chan SA, Singh D, Wei N, Phoo WW, Luo D, Lescar J, Vasudevan SG (2017): "NS3 helicase from dengue virus specifically recognizes viral RNA sequence to ensure optimal replication." Nucleic Acids Res., 45, 12904-12920. doi: 10.1093/nar/gkx1127.
- Abstract
- The protein-RNA interactions within the flavivirus replication complex (RC) are not fully understood. Our structure of dengue virus NS3 adenosine triphosphatase (ATPase)/helicase bound to the conserved 5' genomic RNA 5'-AGUUGUUAGUCU-3' reveals that D290 and R538 make specific interactions with G2 and G5 bases respectively. We show that single-stranded 12-mer RNA stimulates ATPase activity of NS3, however the presence of G2 and G5 leads to significantly higher activation. D290 is adjacent to the DEXH motif found in SF2 helicases like NS3 and interacts with R387, forming a molecular switch that activates the ATPase site upon RNA binding. Our structure guided mutagenesis revealed that disruption of D290-R387 interaction increases basal ATPase activity presumably as a result of higher conformational flexibility of the ATPase active site. Mutational studies also showed R538 plays a critical role in RNA interactions affecting translocation of viral RNA through dynamic interactions with bases at positions 4 and 5 of the ssRNA. Restriction of backbone flexibility around R538 through mutation of G540 to proline abolishes virus replication, indicating conformational flexibility around residue R538 is necessary for RNA translocation. The functionally critical sequence-specific contacts in NS3 RNA binding groove in subdomain III reveals potentially novel allosteric anti-viral drug targets.