Summary information and primary citation
- PDB-id
- 5xm9; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (3.053 Å)
- Summary
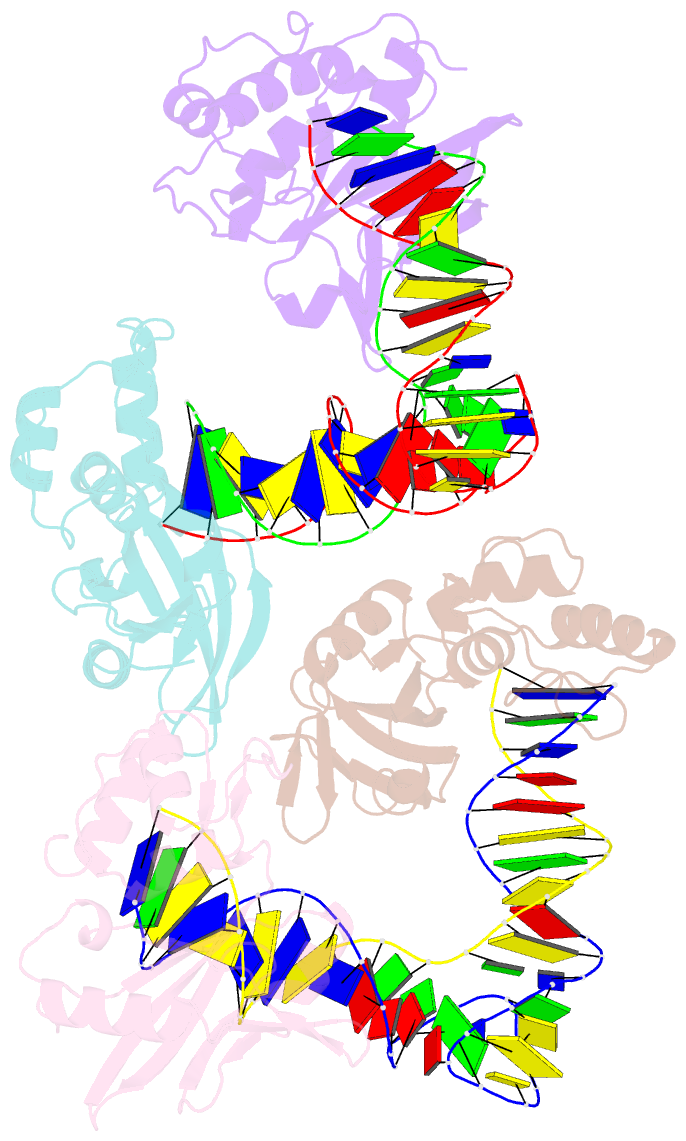
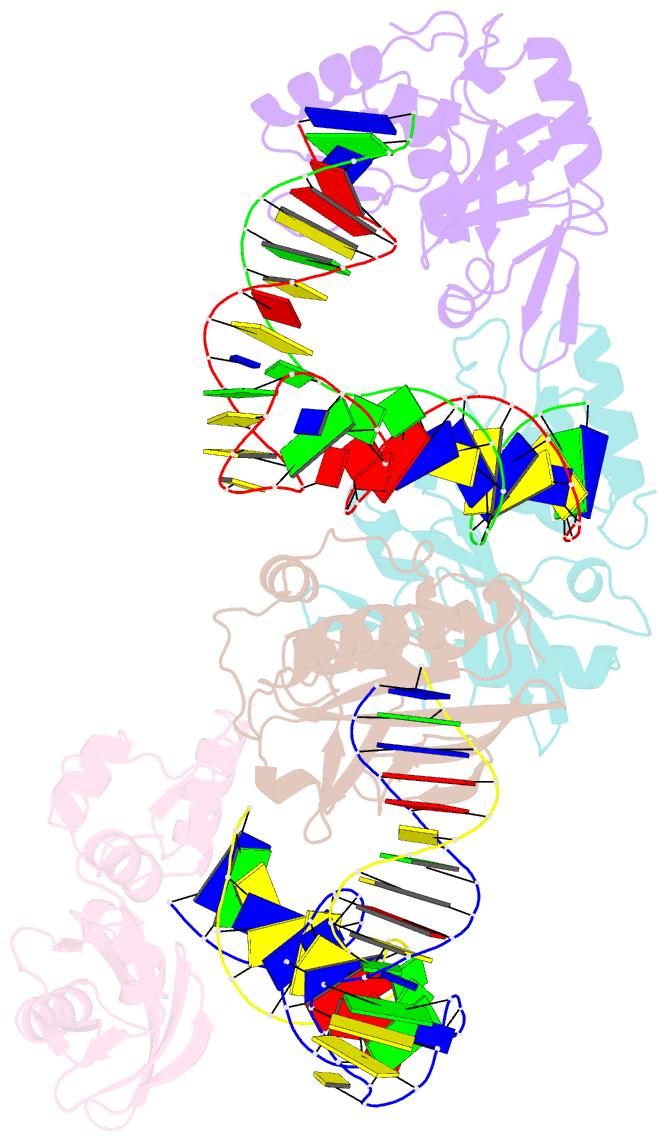
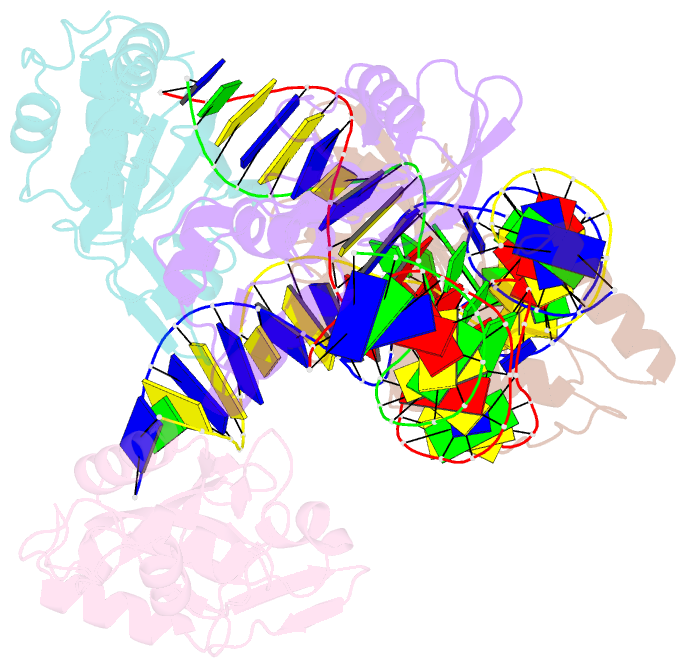
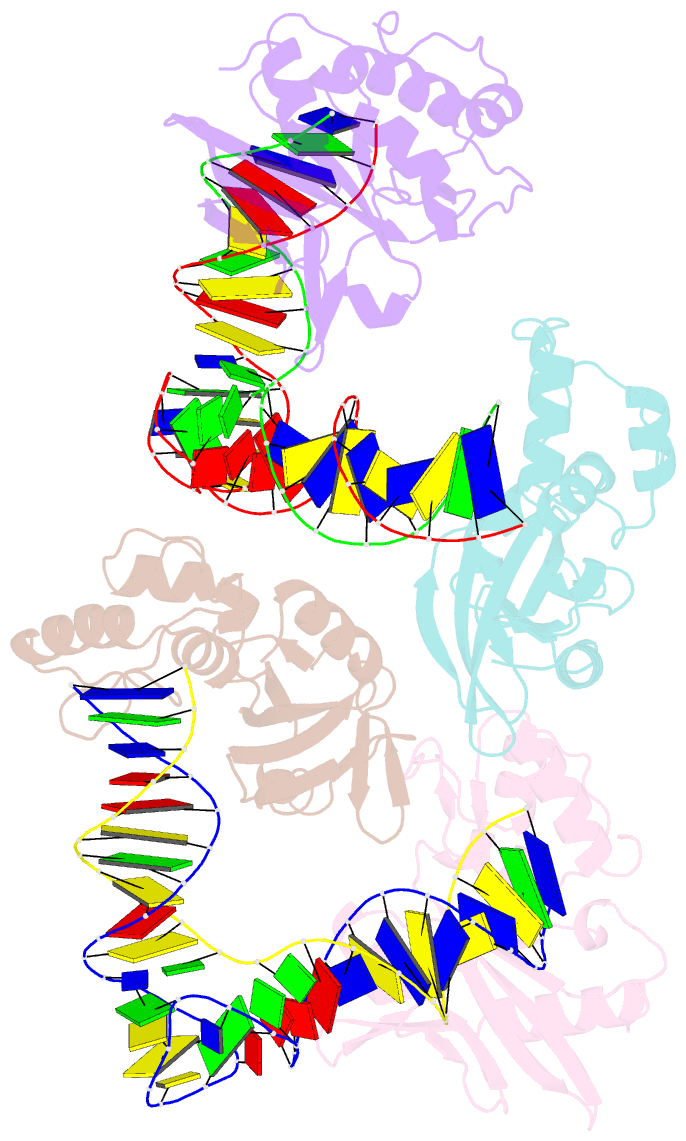
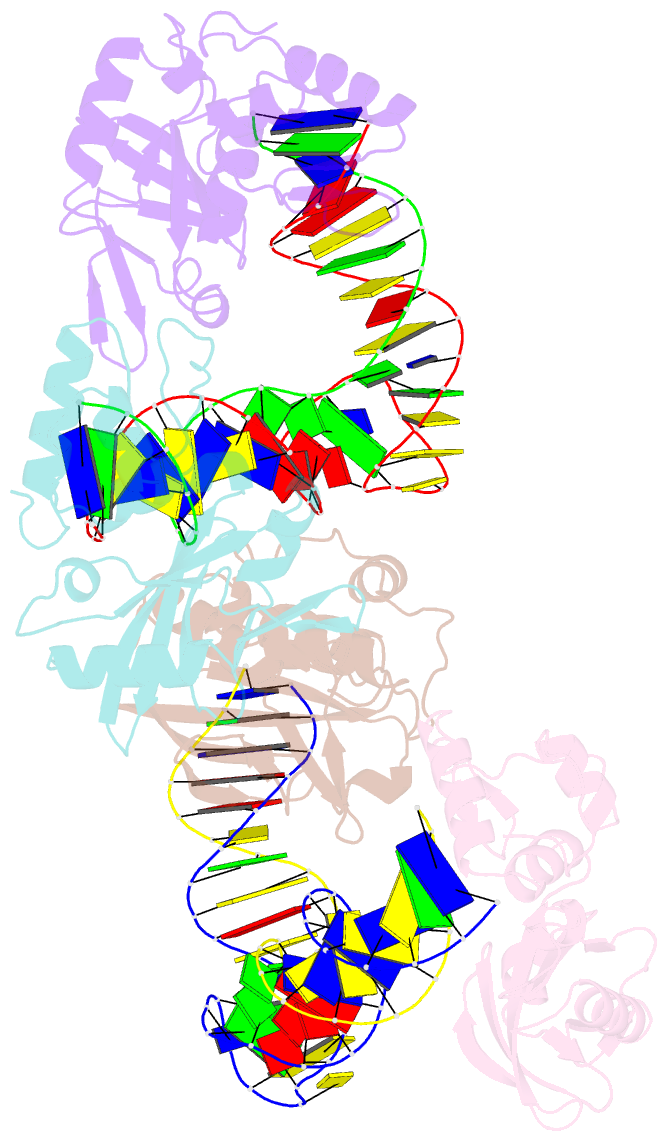
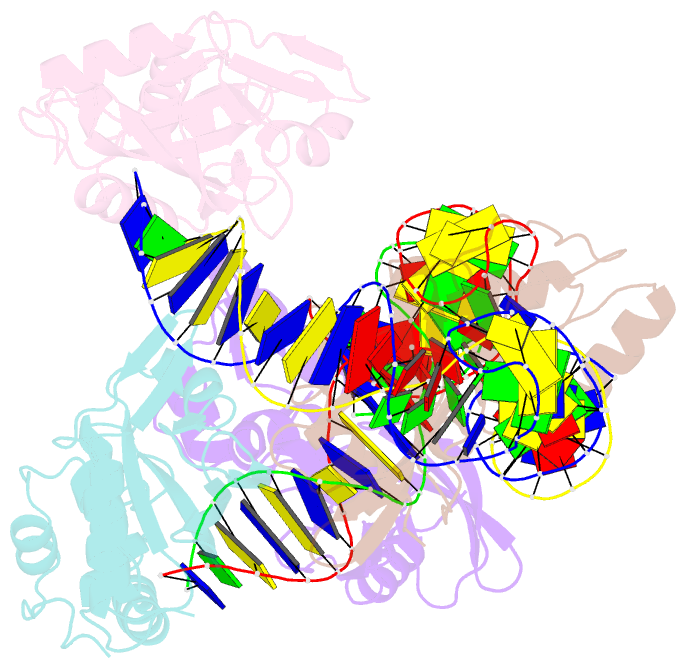
- Crystal structure of asfvpolx in complex with DNA enzyme.
- Reference
- Liu H, Yu X, Chen Y, Zhang J, Wu B, Zheng L, Haruehanroengra P, Wang R, Li S, Lin J, Li J, Sheng J, Huang Z, Ma J, Gan J (2017): "Crystal structure of an RNA-cleaving DNAzyme." Nat Commun, 8, 2006. doi: 10.1038/s41467-017-02203-x.
- Abstract
- In addition to storage of genetic information, DNA can also catalyze various reactions. RNA-cleaving DNAzymes are the catalytic DNAs discovered the earliest, and they can cleave RNAs in a sequence-specific manner. Owing to their great potential in medical therapeutics, virus control, and gene silencing for disease treatments, RNA-cleaving DNAzymes have been extensively studied; however, the mechanistic understandings of their substrate recognition and catalysis remain elusive. Here, we report three catalytic form 8-17 DNAzyme crystal structures. 8-17 DNAzyme adopts a V-shape fold, and the Pb2+ cofactor is bound at the pre-organized pocket. The structures with Pb2+ and the modification at the cleavage site captured the pre-catalytic state of the RNA cleavage reaction, illustrating the unexpected Pb2+-accelerated catalysis, intrinsic tertiary interactions, and molecular kink at the active site. Our studies reveal that DNA is capable of forming a compacted structure and that the functionality-limited bio-polymer can have a novel solution for a functional need in catalysis.