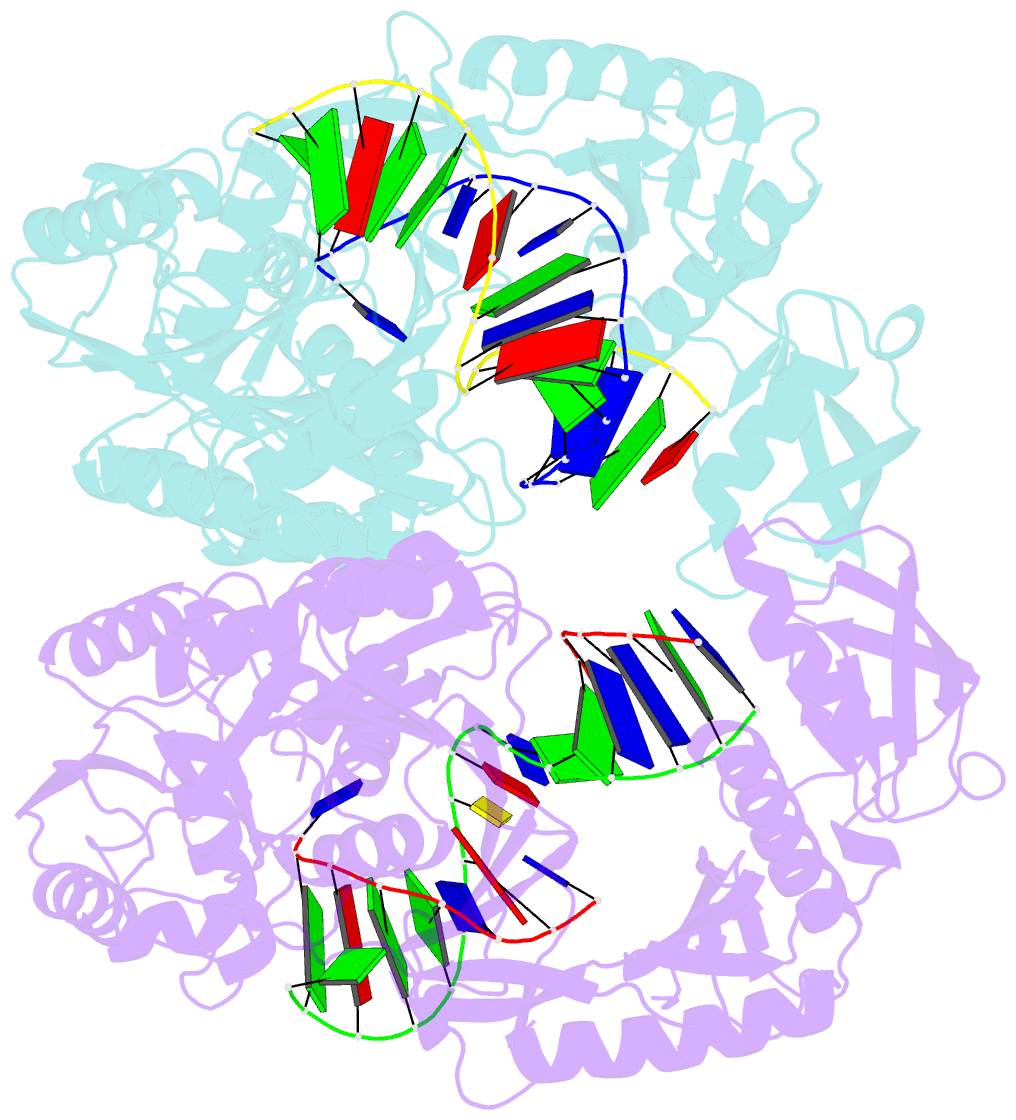
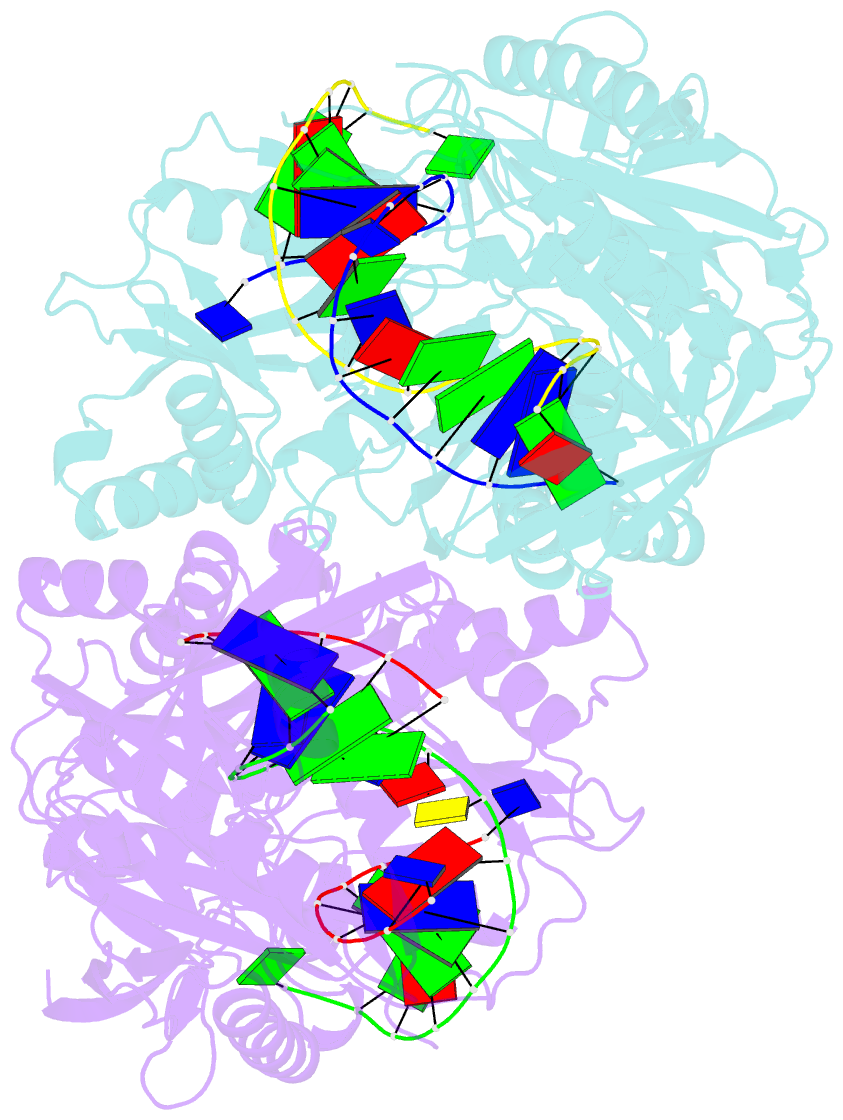
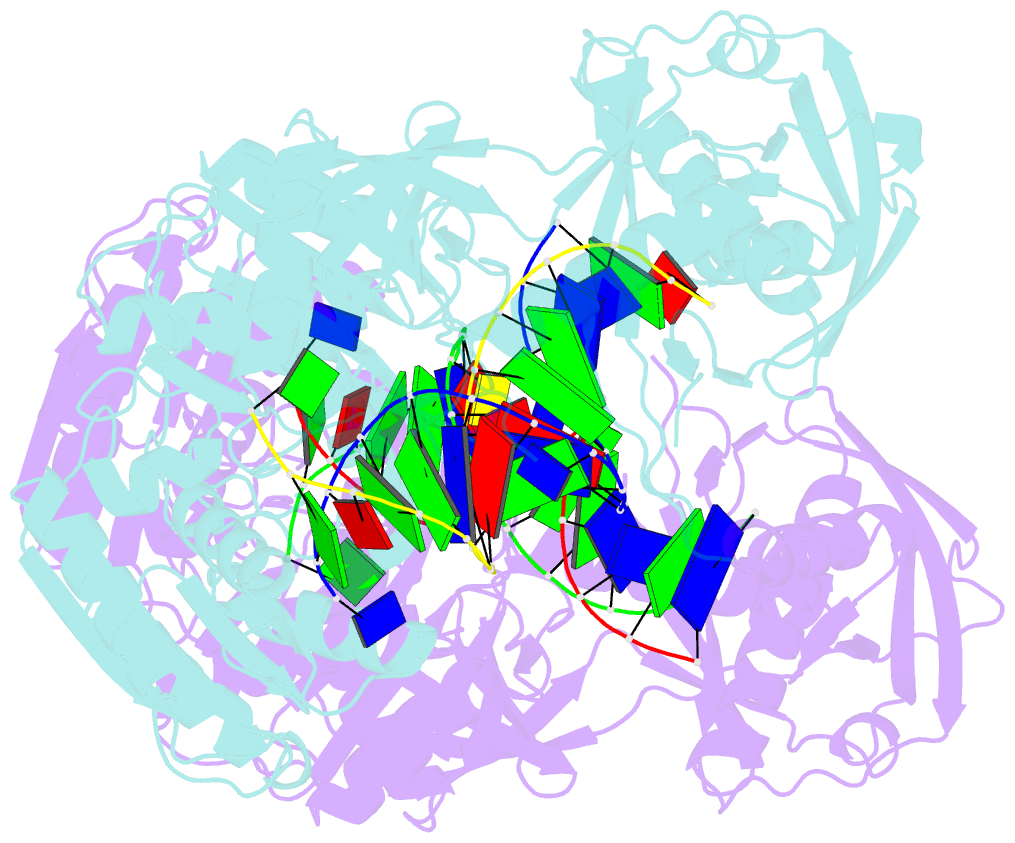
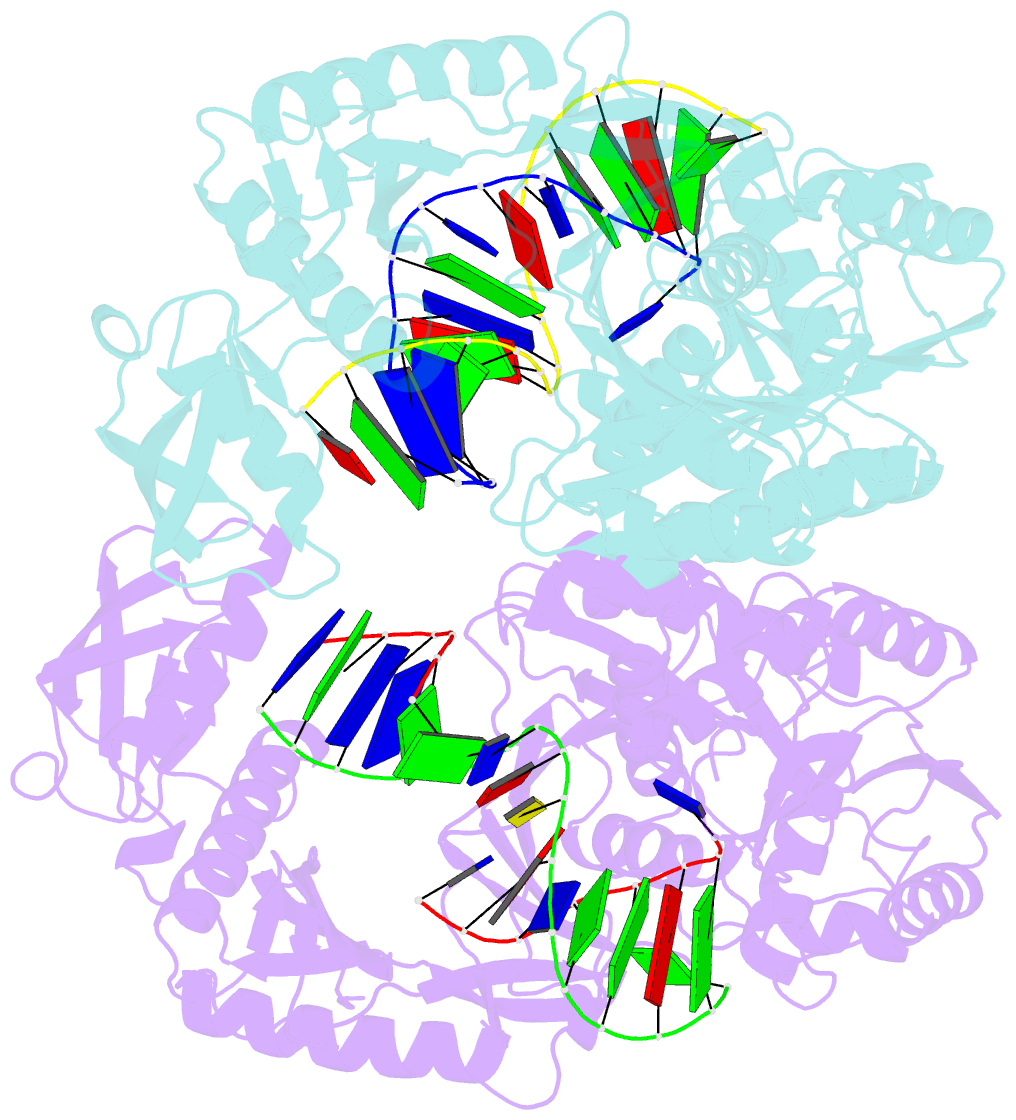
Summary information and primary citation
- PDB-id
- 5xou; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- X-ray (2.63 Å)
- Summary
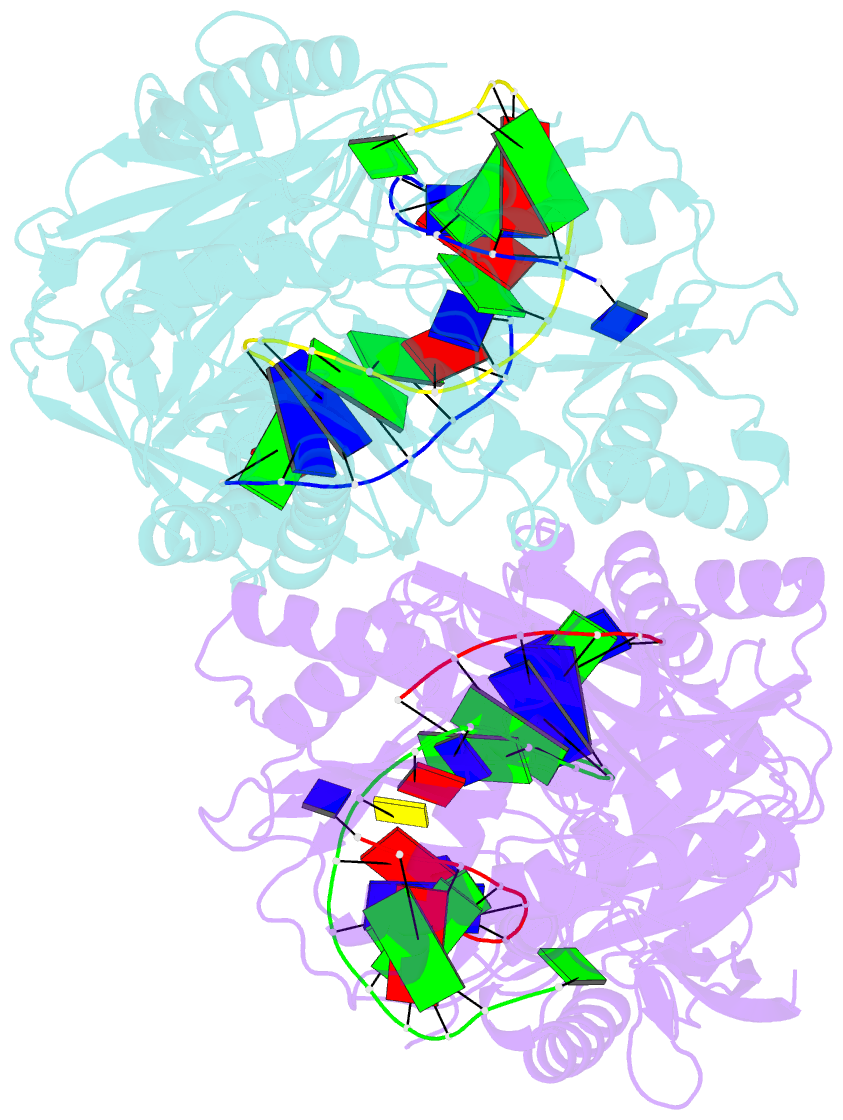
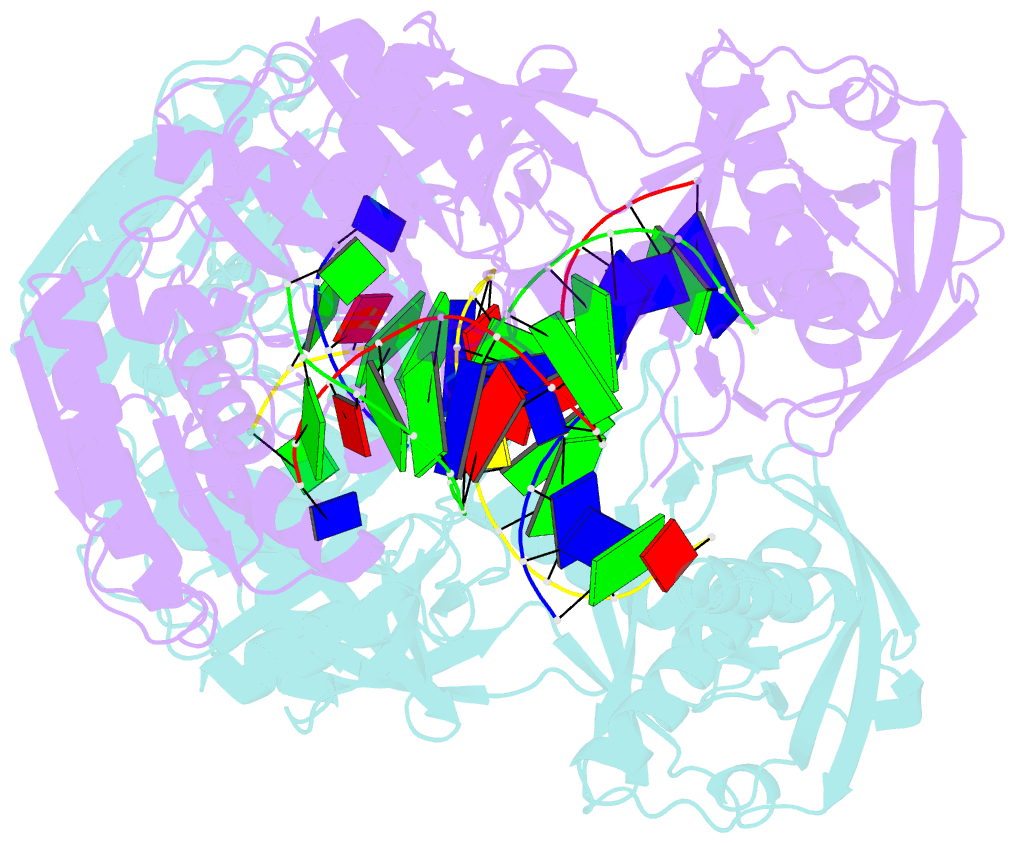
- Crystal structure of t. thermophilus argonaute protein complexed with a bulge 7t8 on the guide strand
- Reference
- Sheng G, Gogakos T, Wang J, Zhao H, Serganov A, Juranek S, Tuschl T, Patel DJ, Wang Y (2017): "Structure/cleavage-based insights into helical perturbations at bulge sites within T. thermophilus Argonaute silencing complexes." Nucleic Acids Res., 45, 9149-9163. doi: 10.1093/nar/gkx547.
- Abstract
- We have undertaken a systematic structural study of Thermus thermophilus Argonaute (TtAgo) ternary complexes containing single-base bulges positioned either within the seed segment of the guide or target strands and at the cleavage site. Our studies establish that single-base bulges 7T8, 5A6 and 4A5 on the guide strand are stacked-into the duplex, with conformational changes localized to the bulge site, thereby having minimal impact on the cleavage site. By contrast, single-base bulges 6'U7' and 6'A7' on the target strand are looped-out of the duplex, with the resulting conformational transitions shifting the cleavable phosphate by one step. We observe a stable alignment for the looped-out 6'N7' bulge base, which stacks on the unpaired first base of the guide strand, with the looped-out alignment facilitated by weakened Watson-Crick and reversed non-canonical flanking pairs. These structural studies are complemented by cleavage assays that independently monitor the impact of bulges on TtAgo-mediated cleavage reaction.