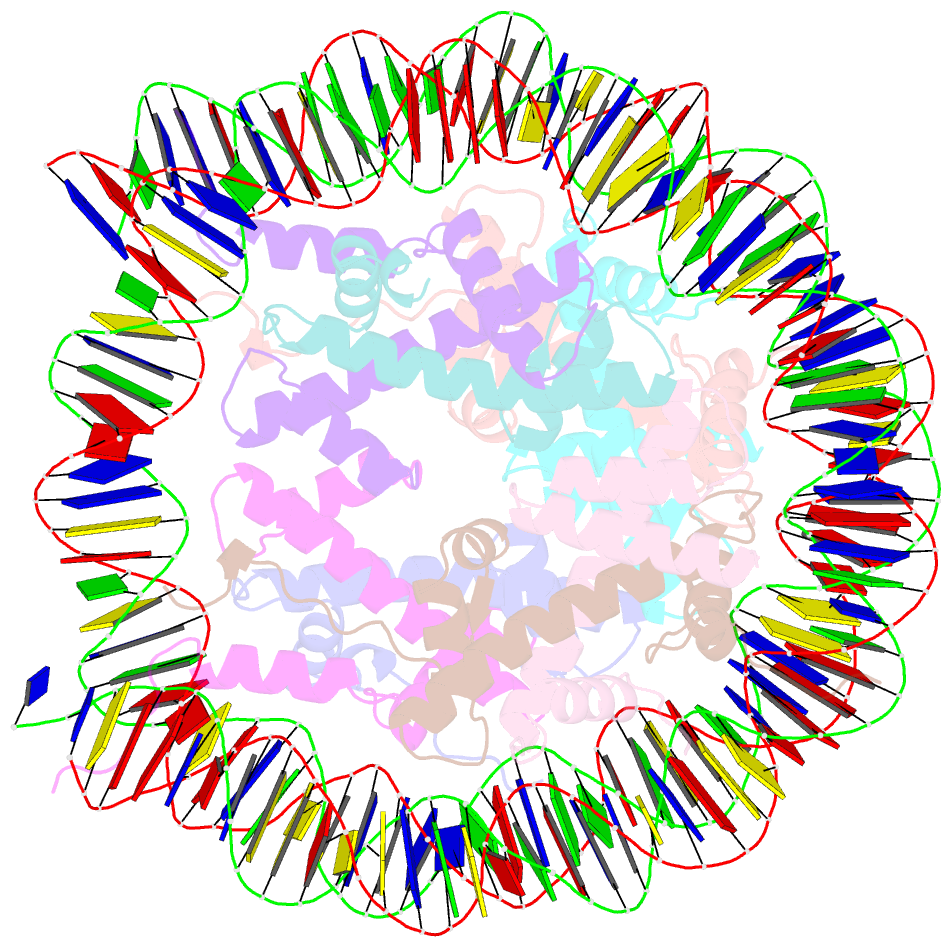
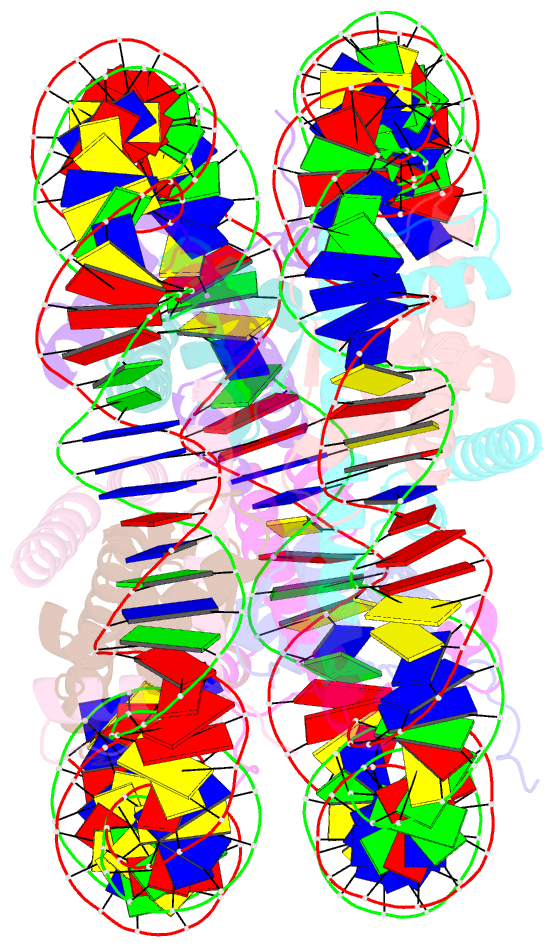
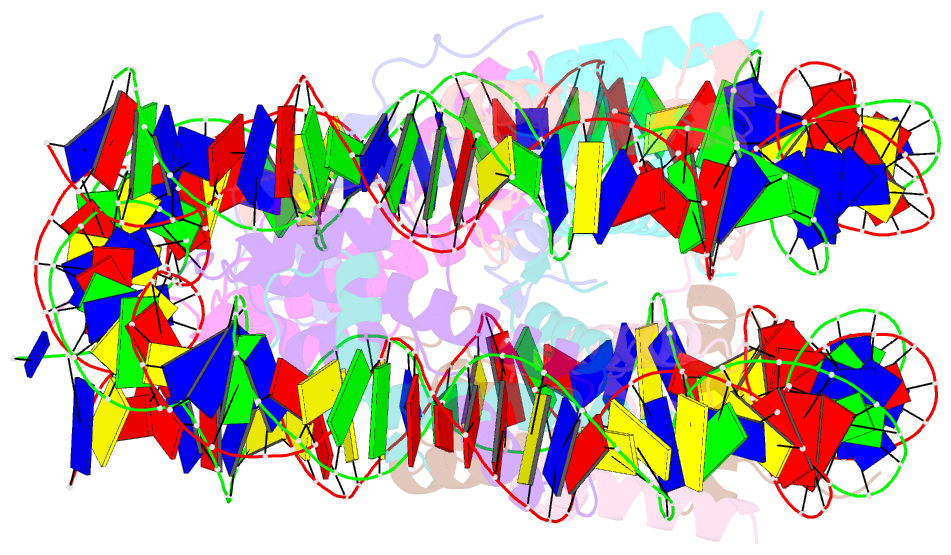
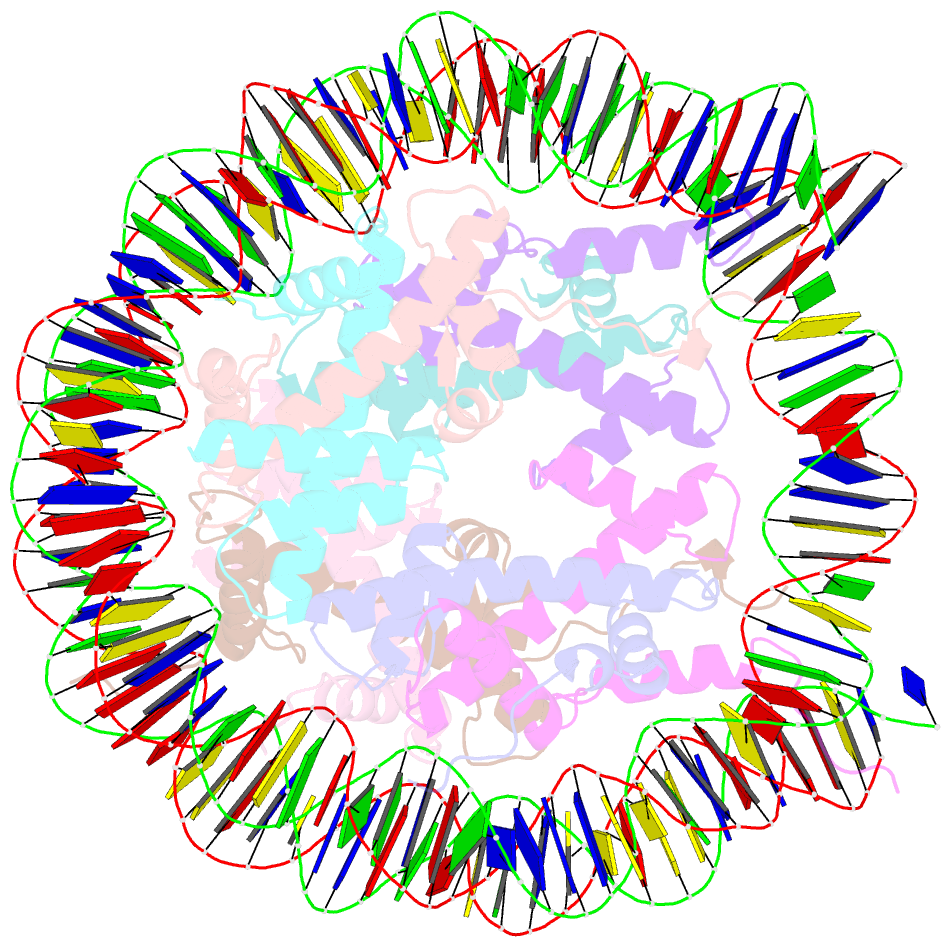
Summary information and primary citation
- PDB-id
- 5y0c; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.087 Å)
- Summary
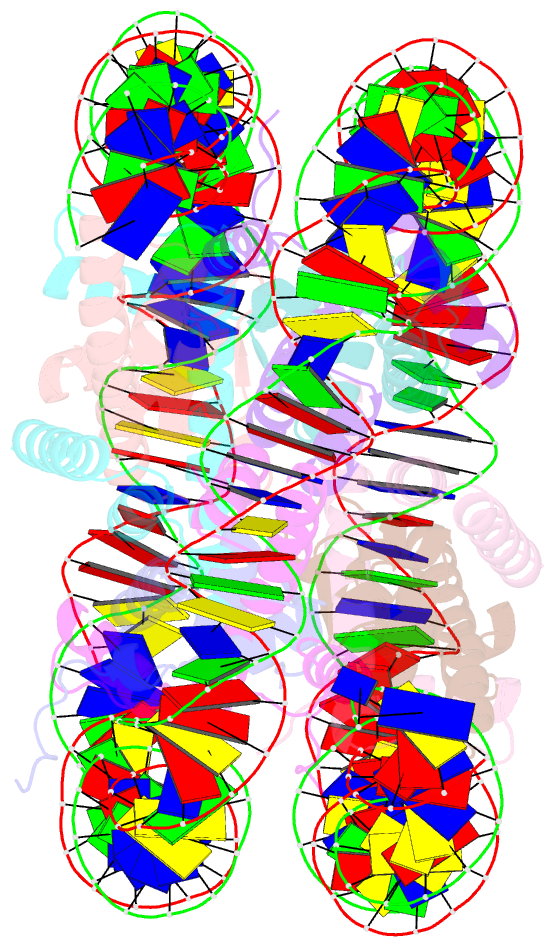
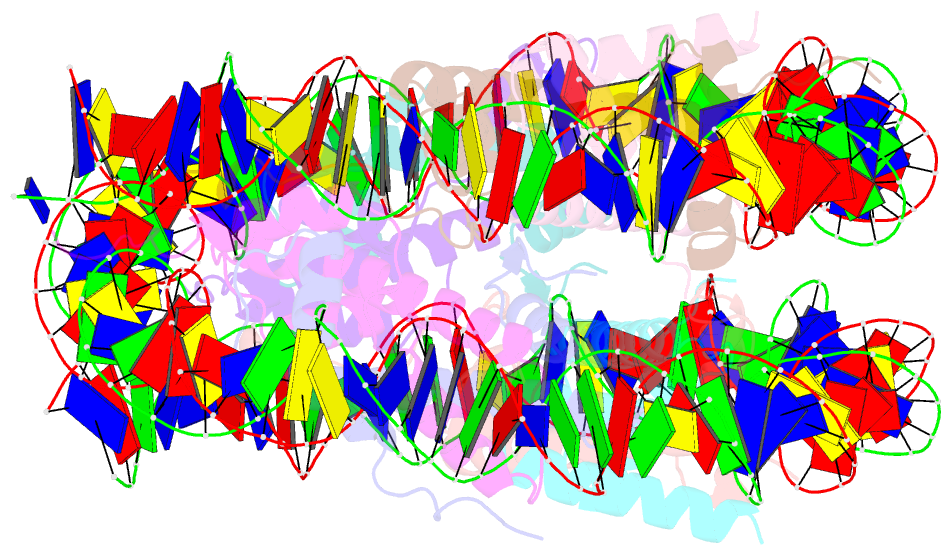
- Crystal structure of the human nucleosome at 2.09 angstrom resolution
- Reference
- Arimura Y, Ikura M, Fujita R, Noda M, Kobayashi W, Horikoshi N, Sun J, Shi L, Kusakabe M, Harata M, Ohkawa Y, Tashiro S, Kimura H, Ikura T, Kurumizaka H (2018): "Cancer-associated mutations of histones H2B, H3.1 and H2A.Z.1 affect the structure and stability of the nucleosome." Nucleic Acids Res., 46, 10007-10018. doi: 10.1093/nar/gky661.
- Abstract
- Mutations of the Glu76 residue of canonical histone H2B are frequently found in cancer cells. However, it is quite mysterious how a single amino acid substitution in one of the multiple H2B genes affects cell fate. Here we found that the H2B E76K mutation, in which Glu76 is replaced by Lys (E76K), distorted the interface between H2B and H4 in the nucleosome, as revealed by the crystal structure and induced nucleosome instability in vivo and in vitro. Exogenous production of the H2B E76K mutant robustly enhanced the colony formation ability of the expressing cells, indicating that the H2B E76K mutant has the potential to promote oncogenic transformation in the presence of wild-type H2B. We found that other cancer-associated mutations of histones, H3.1 E97K and H2A.Z.1 R80C, also induced nucleosome instability. Interestingly, like the H2B E76K mutant, the H3.1 E97K mutant was minimally incorporated into chromatin in cells, but it enhanced the colony formation ability. In contrast, the H2A.Z.1 R80C mutant was incorporated into chromatin in cells, and had minor effects on the colony formation ability of the cells. These characteristics of histones with cancer-associated mutations may provide important information toward understanding how the mutations promote cancer progression.