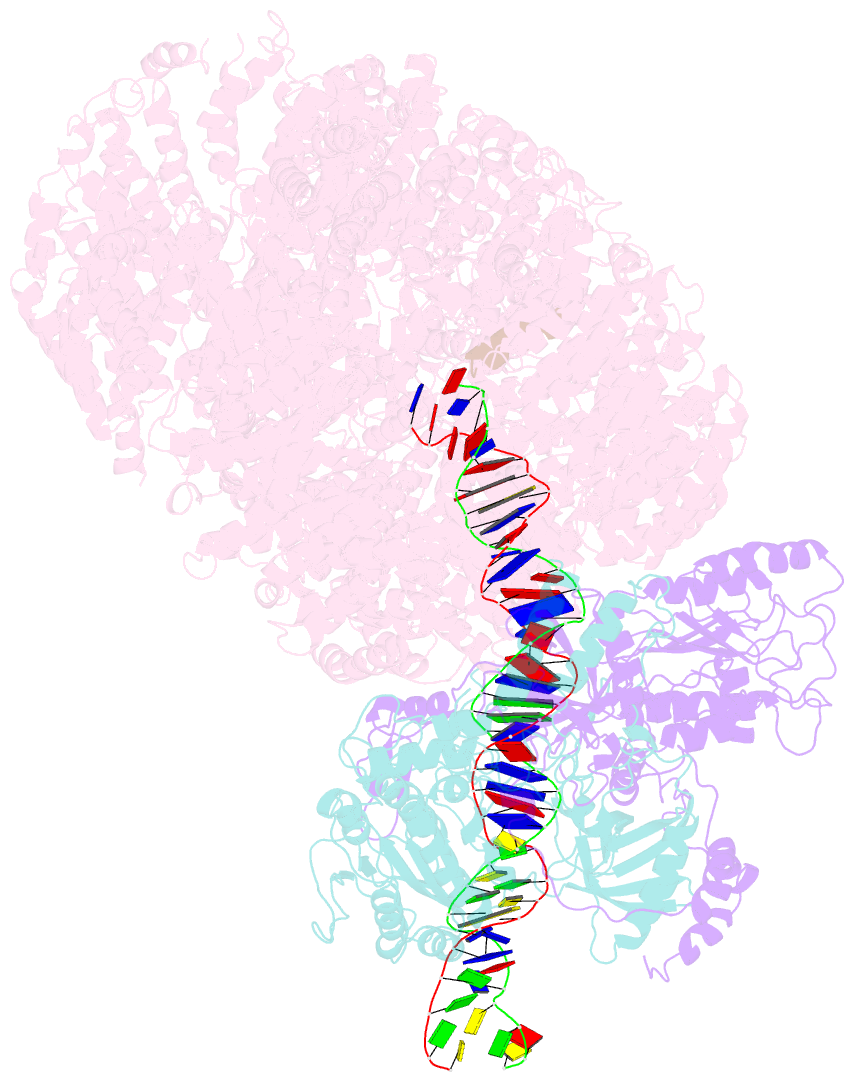
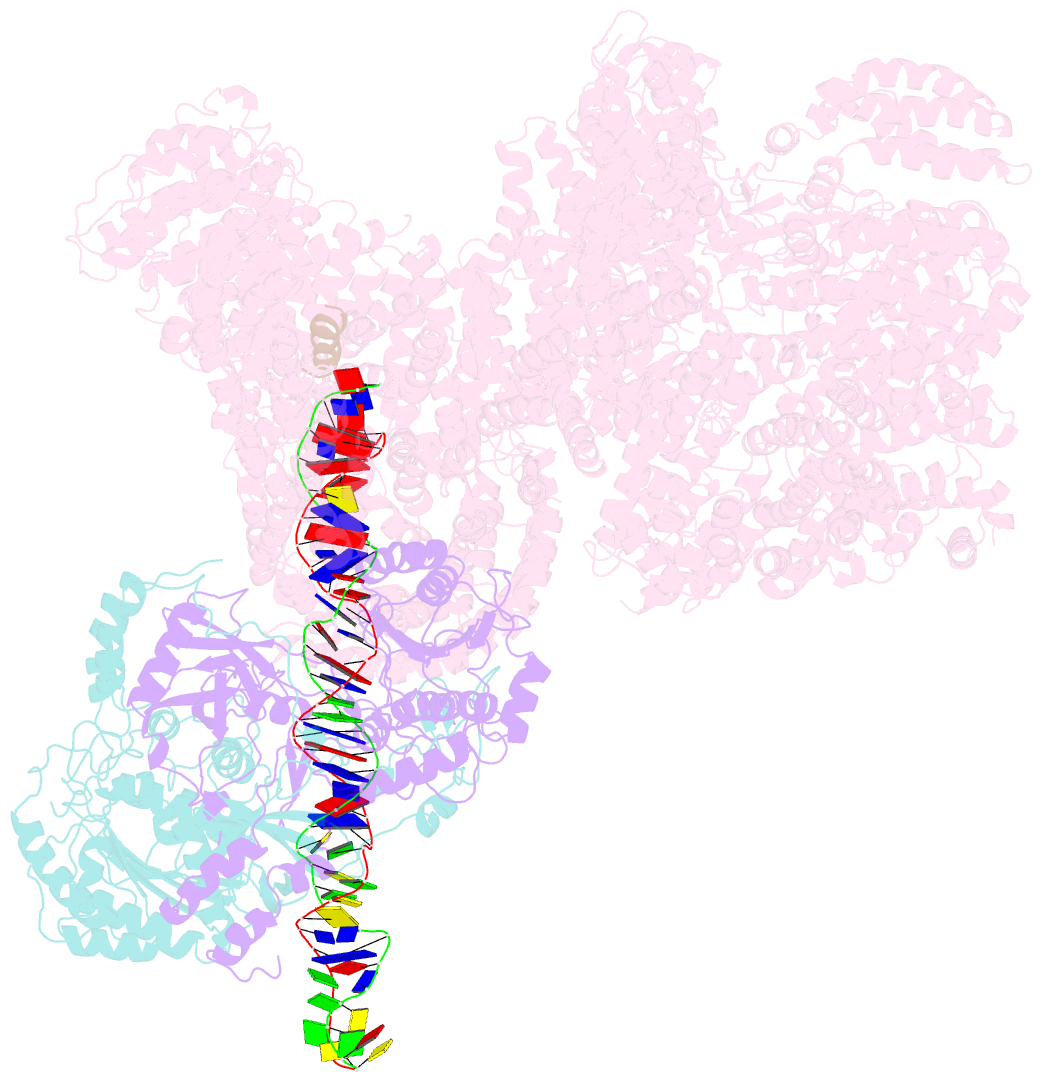
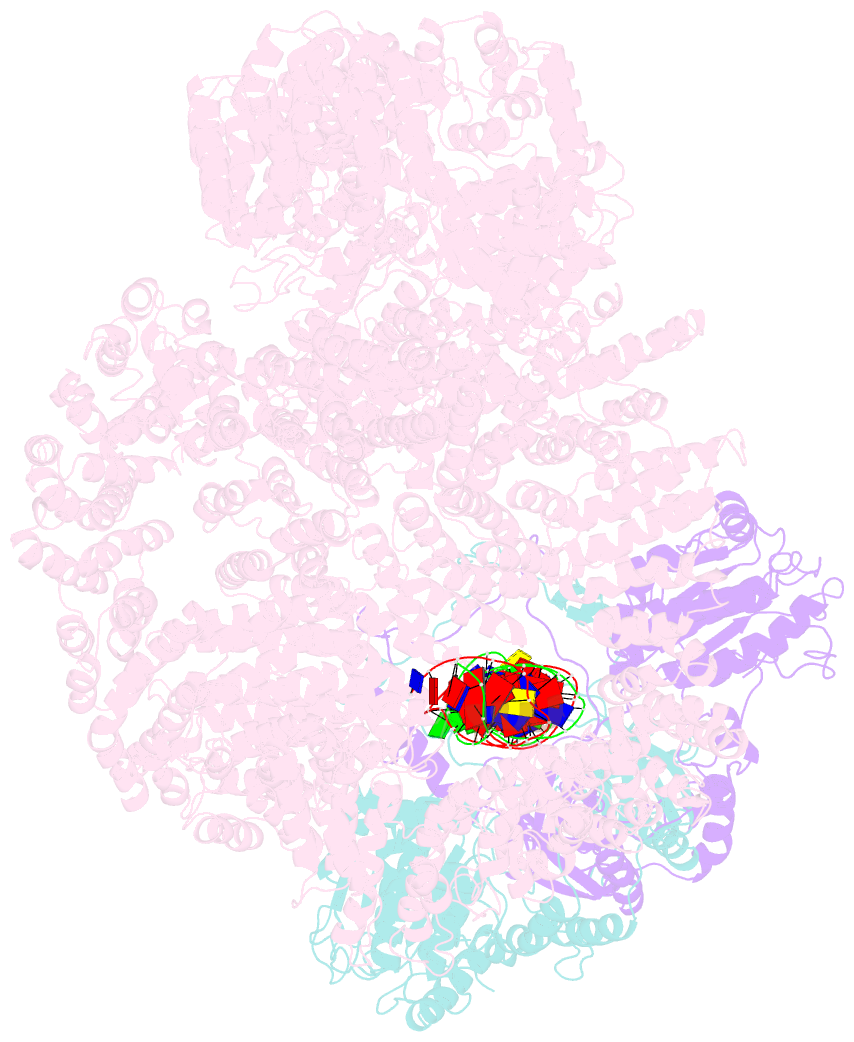
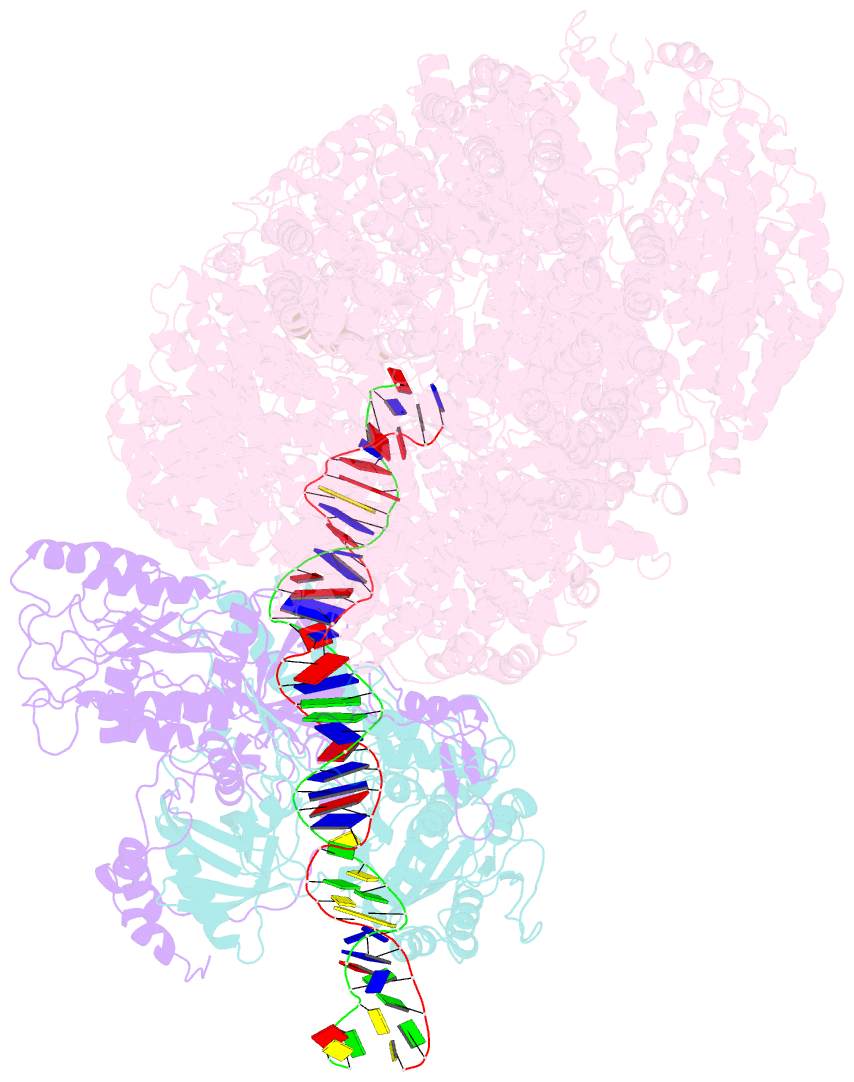
Summary information and primary citation
- PDB-id
- 5y3r; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- cryo-EM (6.6 Å)
- Summary
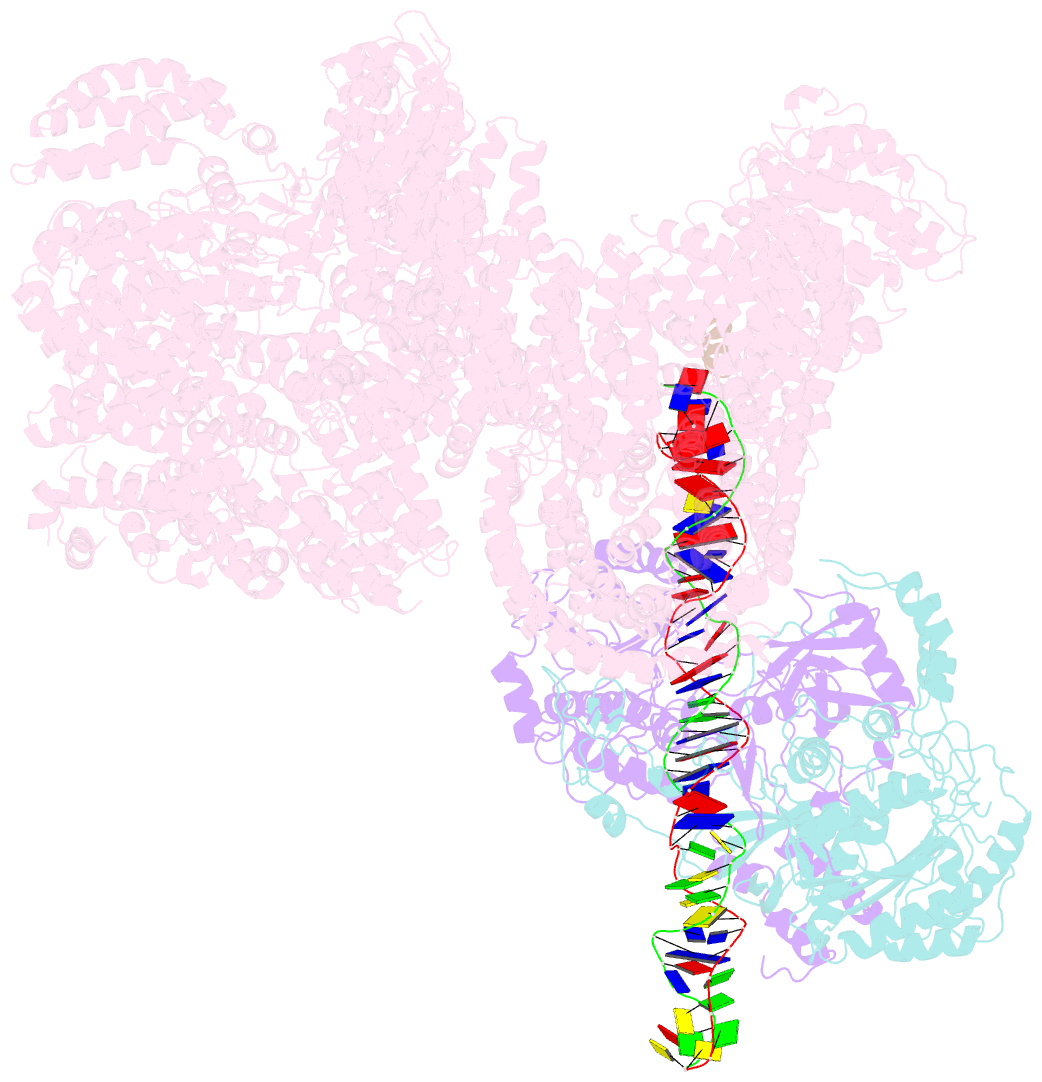
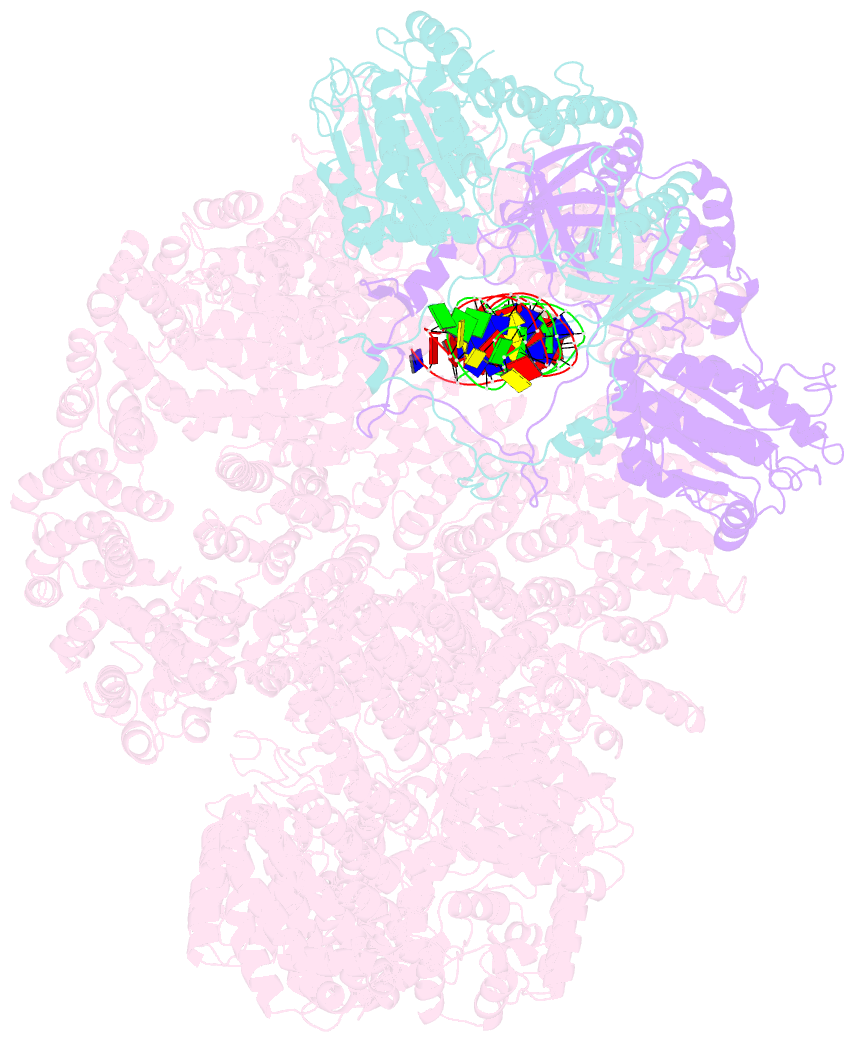
- cryo-EM structure of human DNA-pk holoenzyme
- Reference
- Yin X, Liu M, Tian Y, Wang J, Xu Y (2017): "Cryo-EM structure of human DNA-PK holoenzyme." Cell Res., 27, 1341-1350. doi: 10.1038/cr.2017.110.
- Abstract
- DNA-dependent protein kinase (DNA-PK) is a serine/threonine protein kinase complex composed of a catalytic subunit (DNA-PKcs) and KU70/80 heterodimer bound to DNA. DNA-PK holoenzyme plays a critical role in non-homologous end joining (NHEJ), the major DNA repair pathway. Here, we determined cryo-electron microscopy structure of human DNA-PK holoenzyme at 6.6 Å resolution. In the complex structure, DNA-PKcs, KU70, KU80 and DNA duplex form a 650-kDa heterotetramer with 1:1:1:1 stoichiometry. The N-terminal α-solenoid (∼2 800 residues) of DNA-PKcs adopts a double-ring fold and connects the catalytic core domain of DNA-PKcs and KU70/80-DNA. DNA-PKcs and KU70/80 together form a DNA-binding tunnel, which cradles ∼30-bp DNA and prevents sliding inward of DNA-PKcs along with DNA duplex, suggesting a mechanism by which the broken DNA end is protected from unnecessary processing. Structural and biochemical analyses indicate that KU70/80 and DNA coordinately induce conformational changes of DNA-PKcs and allosterically stimulate its kinase activity. We propose a model for activation of DNA-PKcs in which allosteric signals are generated upon DNA-PK holoenzyme formation and transmitted to the kinase domain through N-terminal HEAT repeats and FAT domain of DNA-PKcs. Our studies suggest a mechanism for recognition and protection of broken DNA ends and provide a structural basis for understanding the activation of DNA-PKcs and DNA-PK-mediated NHEJ pathway.