Summary information and primary citation
- PDB-id
- 5z4d; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-RNA
- Method
- X-ray (1.803 Å)
- Summary
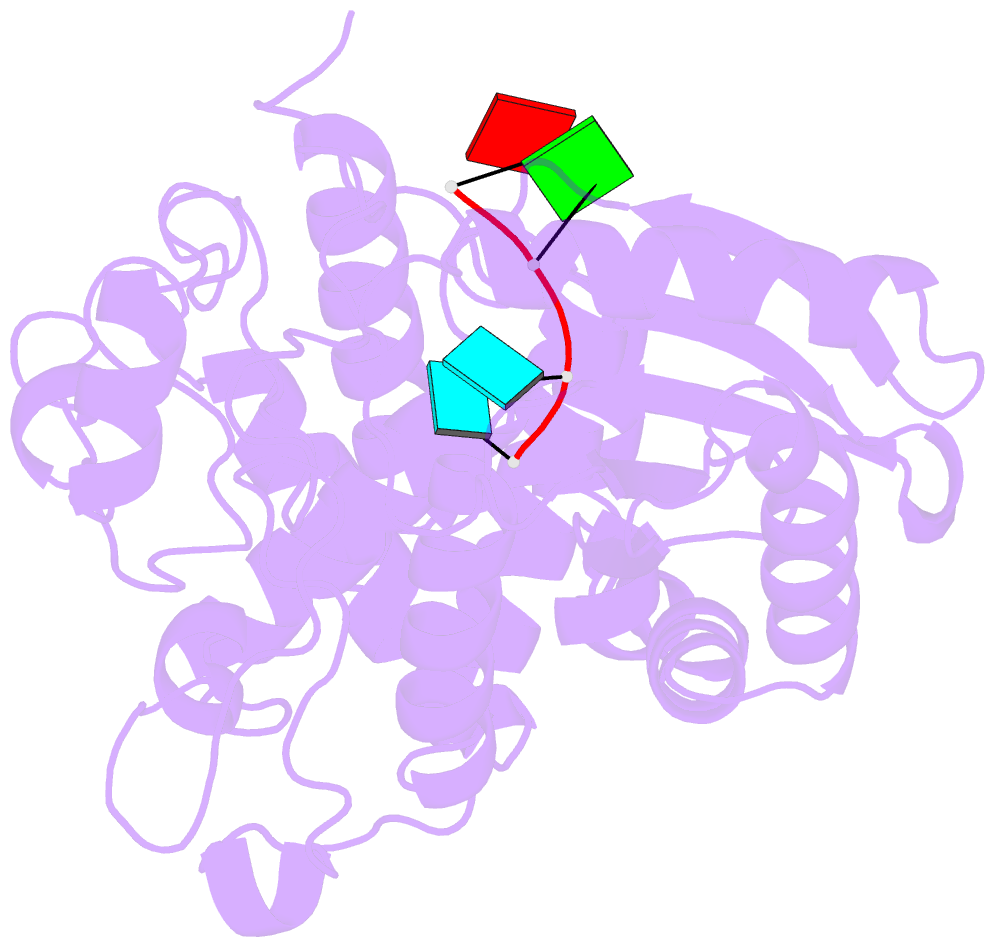
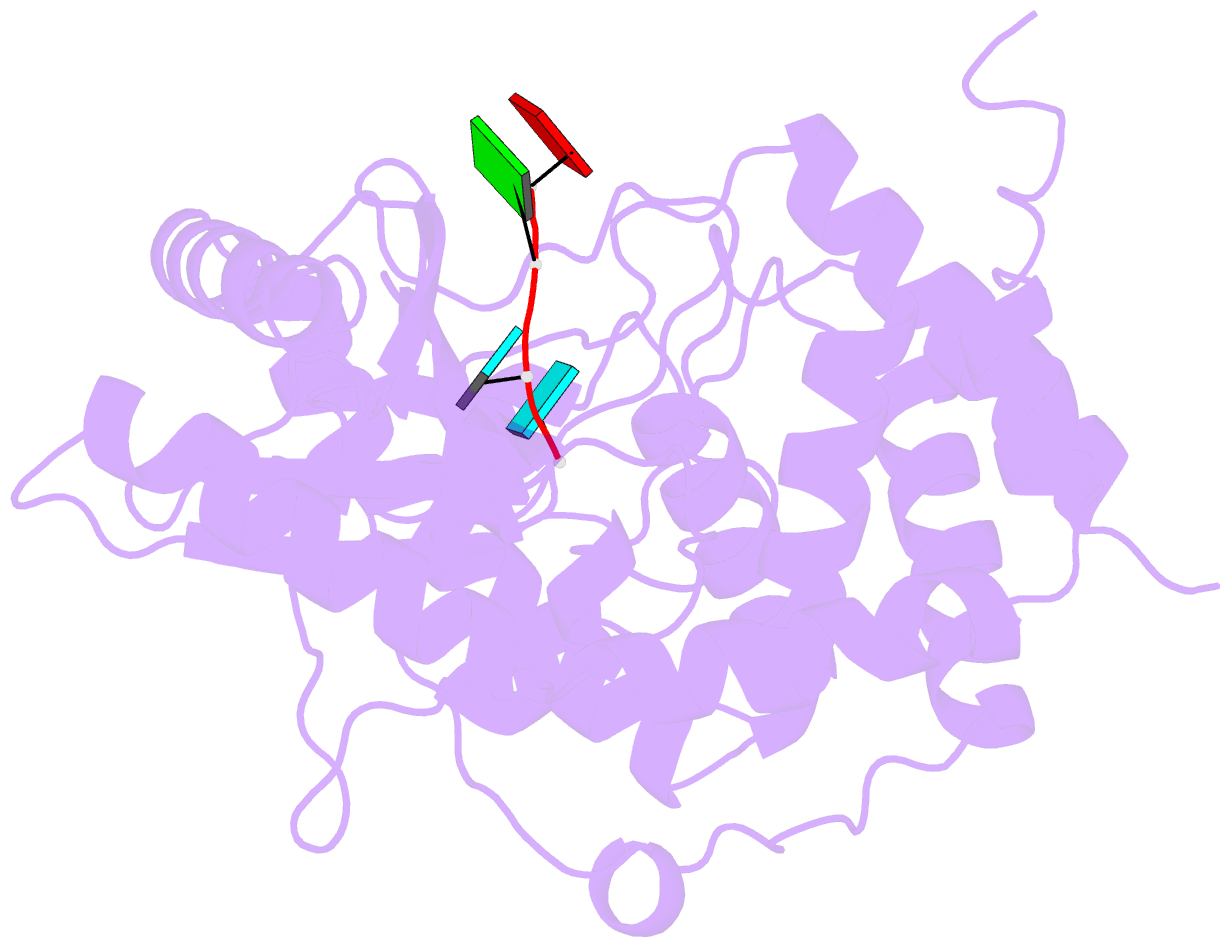
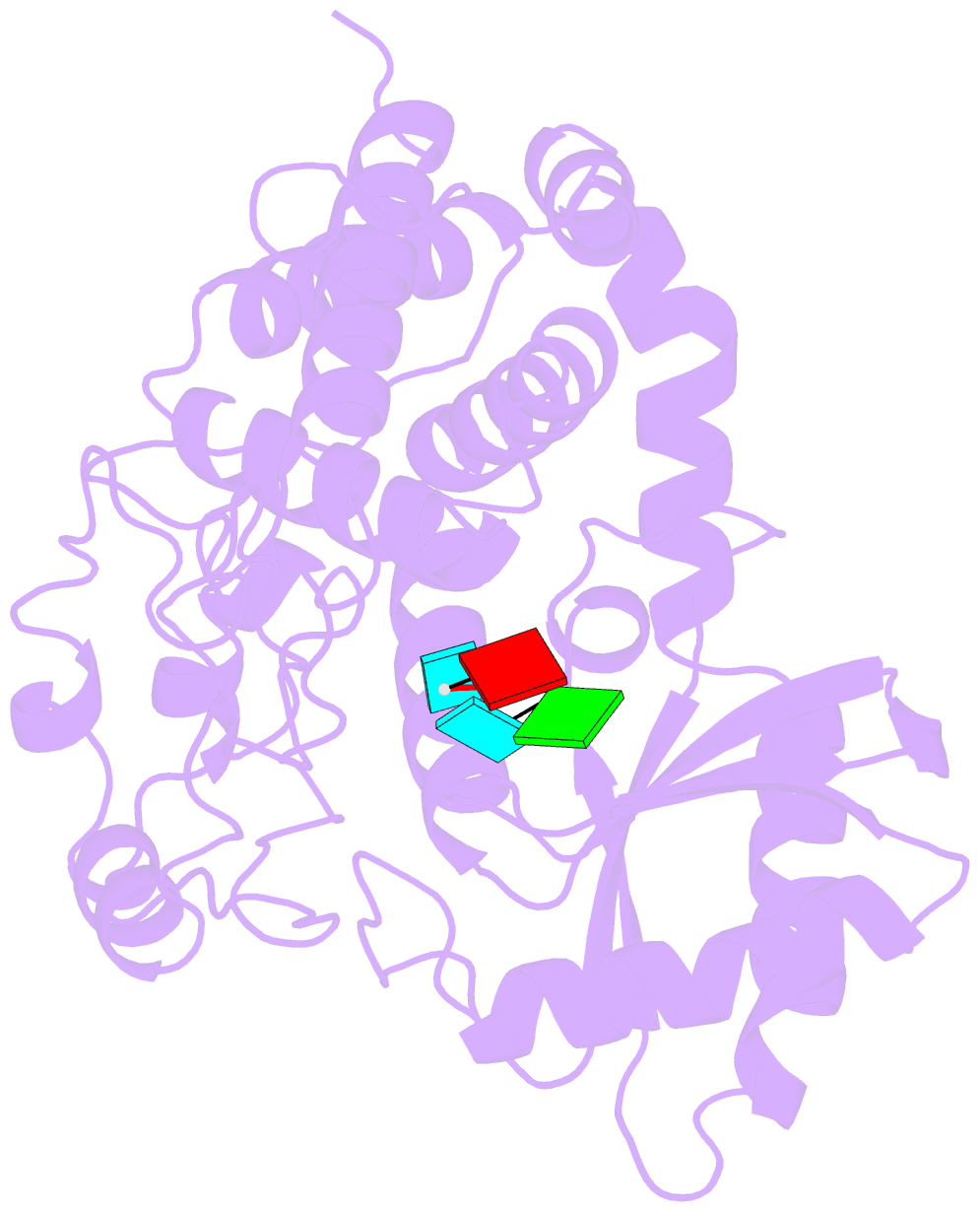
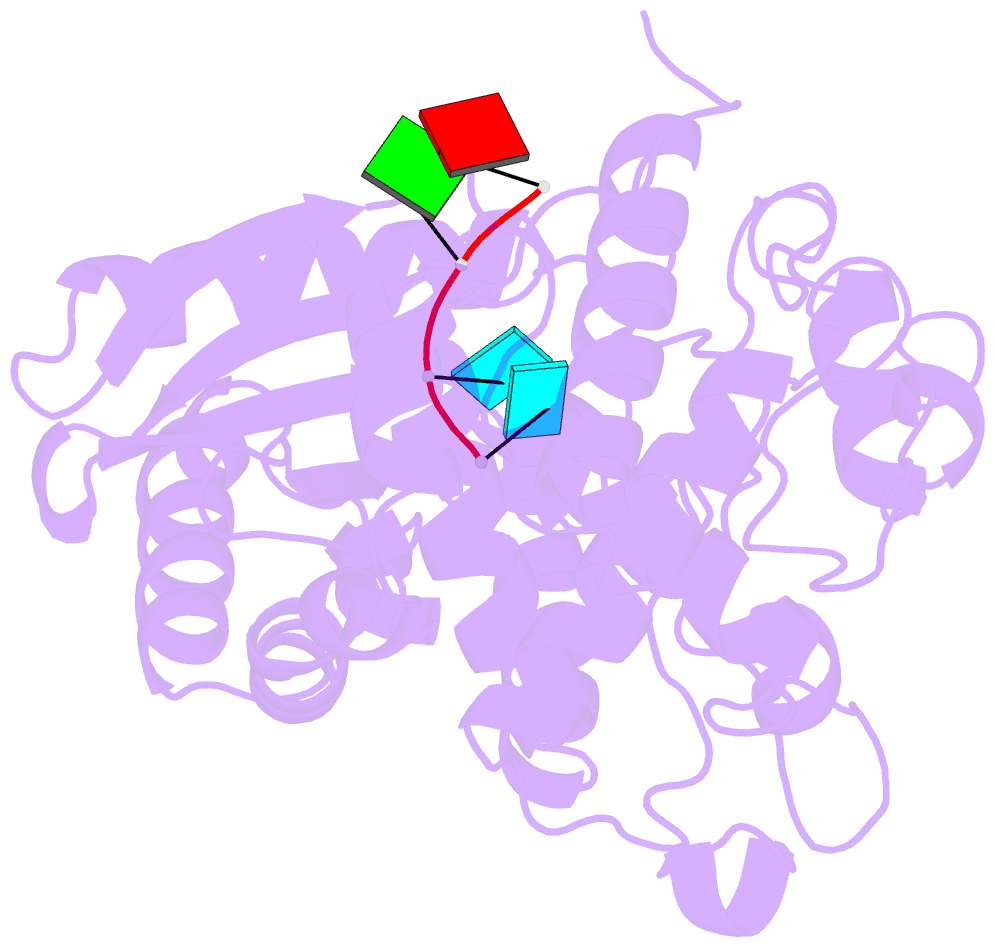
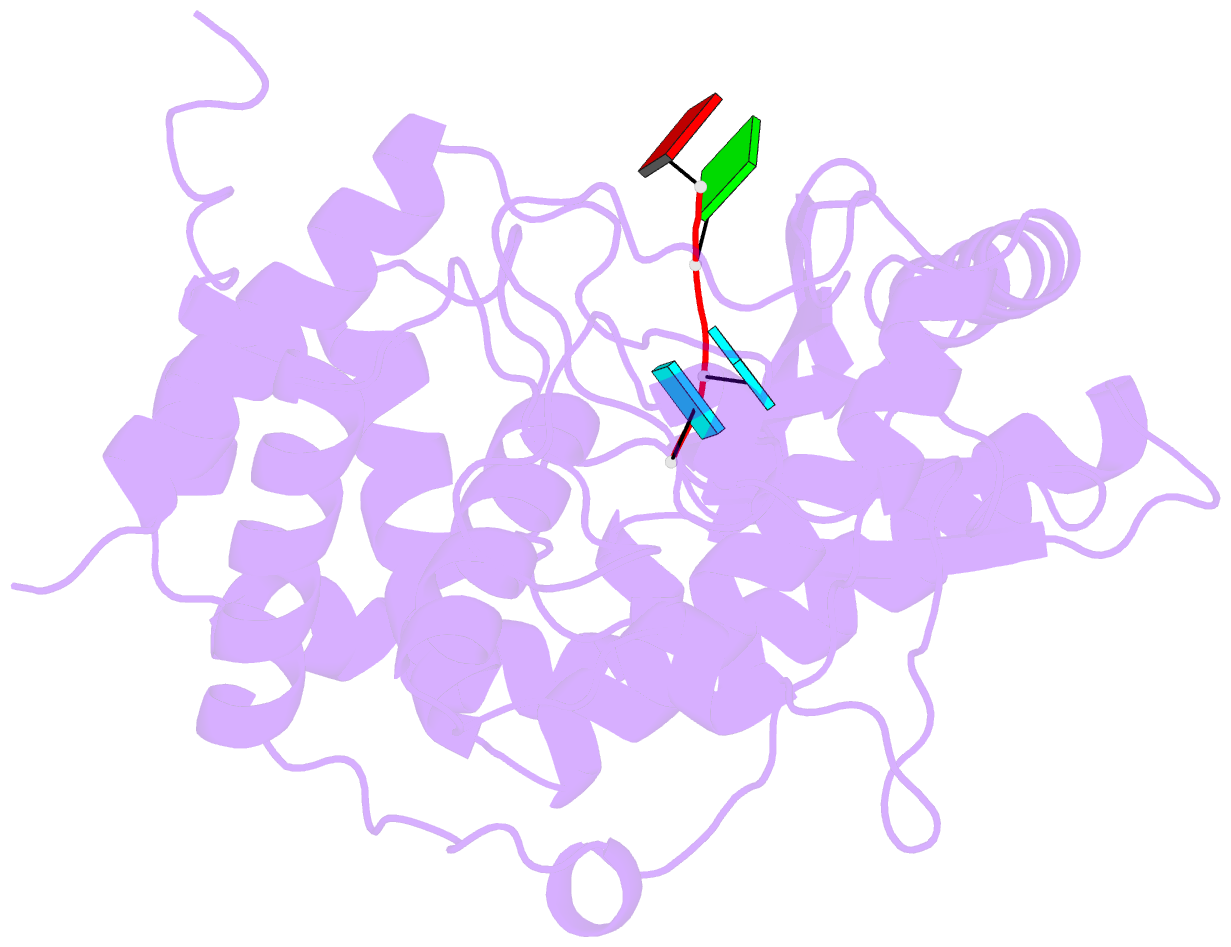
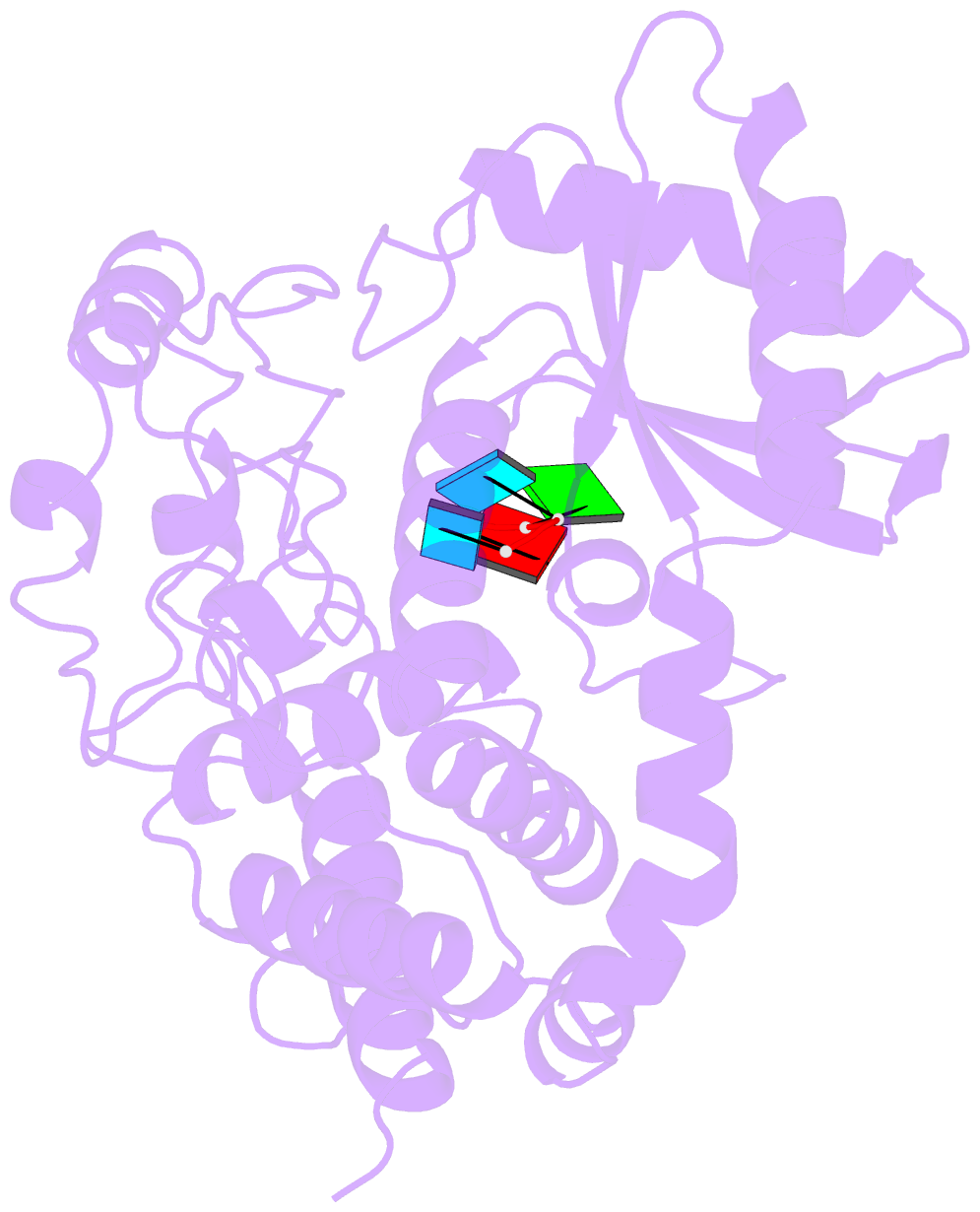
- Structure of tailor in complex with aguu RNA
- Reference
- Cheng L, Li F, Jiang Y, Yu H, Xie C, Shi Y, Gong Q (2019): "Structural insights into a unique preference for 3' terminal guanine of mirtron in Drosophila TUTase tailor." Nucleic Acids Res., 47, 495-508. doi: 10.1093/nar/gky1116.
- Abstract
- Terminal uridylyl transferase (TUTase) is one type of enzyme that modifies RNA molecules by facilitating the post-transcriptional addition of uridyl ribonucleotides to their 3' ends. Recent researches have reported that Drosophila TUTase, Tailor, exhibits an intrinsic preference for RNA substrates ending in 3'G, distinguishing it from any other known TUTases. Through this unique feature, Tailor plays a crucial role as the repressor in the biogenesis pathway of splicing-derived mirtron pre-miRNAs. Here we describe crystal structures of core catalytic domain of Tailor and its complexes with RNA stretches 5'-AGU-3' and 5'-AGUU-3'. We demonstrate that R327 and N347 are two key residues contributing cooperatively to Tailor's preference for 3'G, and R327 may play an extra role in facilitating the extension of polyuridylation chain. We also demonstrate that conformational stability of the exit of RNA-binding groove also contributes significantly to Tailor's activity. Overall, our work reveals useful insights to explain why Drosophila Tailor can preferentially select RNA substrates ending in 3'G and provides important values for further understanding the biological significances of biogenesis pathway of mirtron in flies.