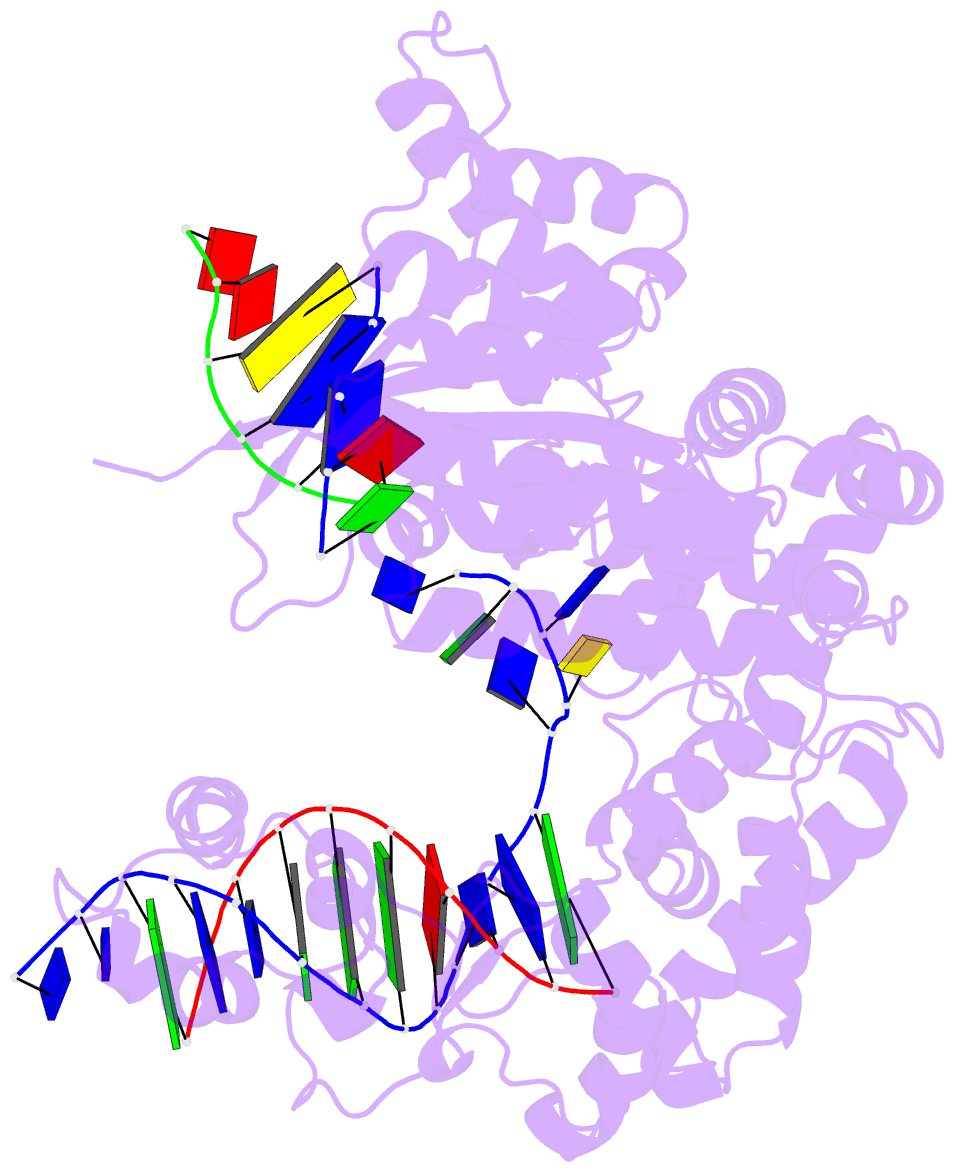
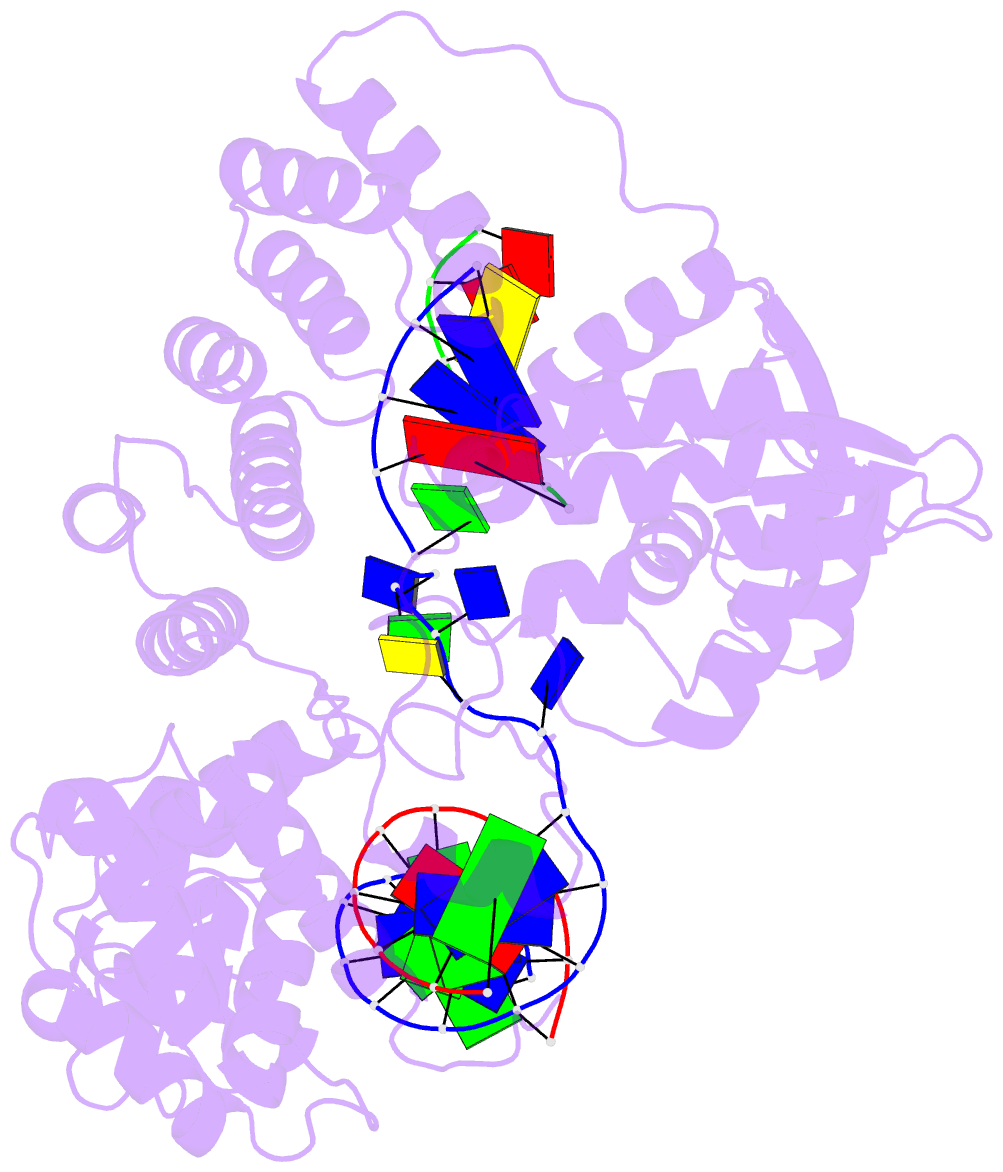
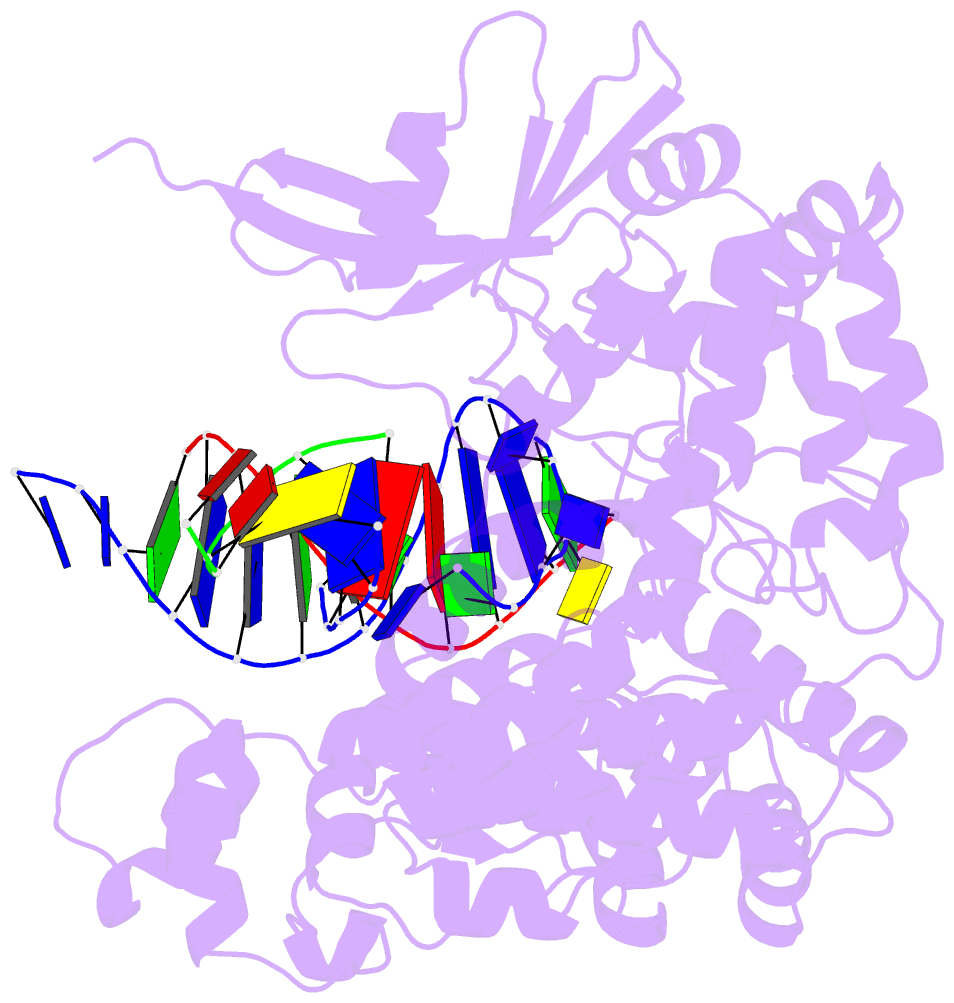
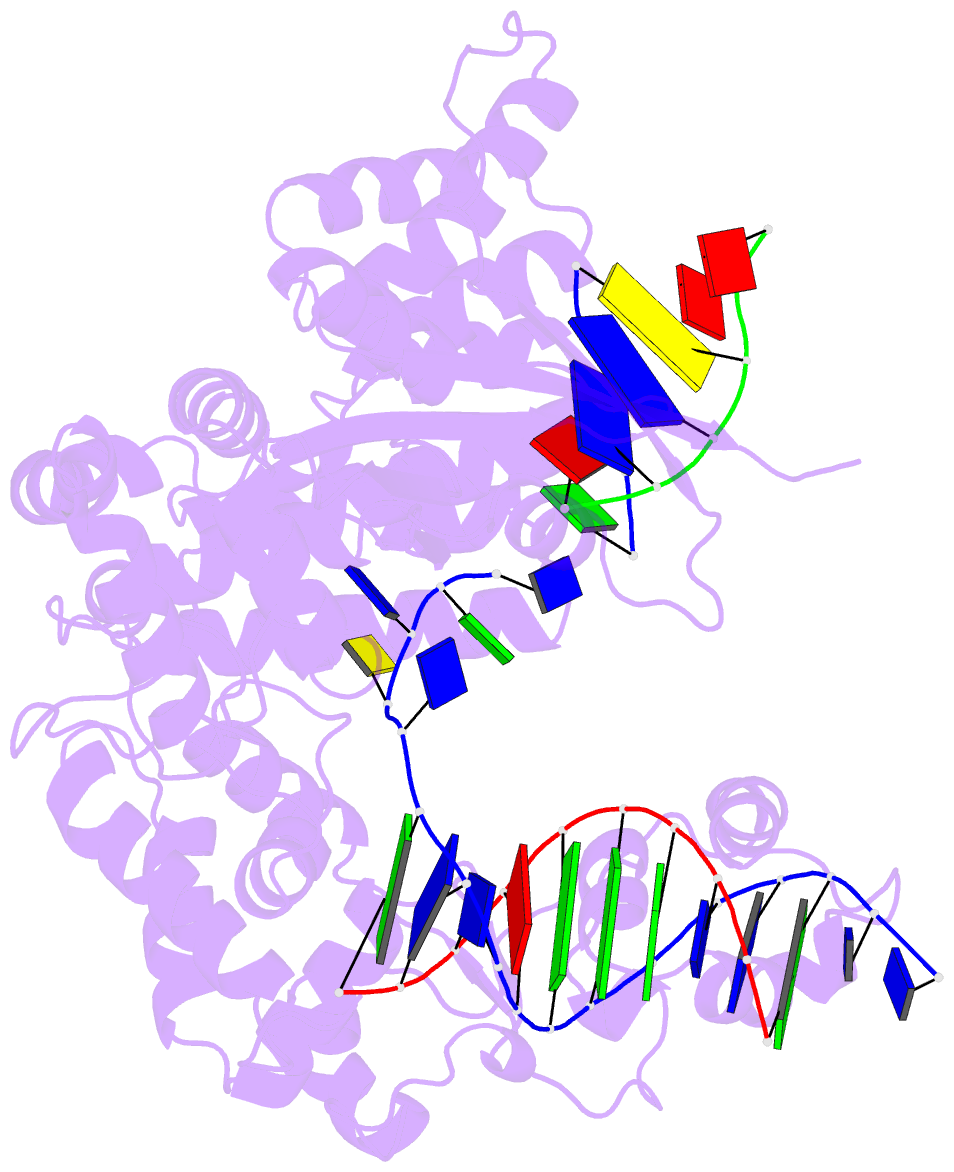
Summary information and primary citation
- PDB-id
- 5z6w; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (3.2 Å)
- Summary
- Crystal structure of pafan1 bound to 2nt 5'flap DNA with gap with manganese
- Reference
- Jin H, Roy U, Lee G, Scharer OD, Cho Y (2018): "Structural mechanism of DNA interstrand cross-link unhooking by the bacterial FAN1 nuclease." J. Biol. Chem., 293, 6482-6496. doi: 10.1074/jbc.RA118.002171.
- Abstract
- DNA interstrand cross-links (ICLs) block the progress of the replication and transcription machineries and can weaken chromosomal stability, resulting in various diseases. FANCD2-FANCI-associated nuclease (FAN1) is a conserved structure-specific nuclease that unhooks DNA ICLs independently of the Fanconi anemia pathway. Recent structural studies have proposed two different mechanistic features for ICL unhooking by human FAN1: a specific basic pocket that recognizes the terminal phosphate of a 1-nucleotide (nt) 5' flap or FAN1 dimerization. Herein, we show that despite lacking these features, Pseudomonas aeruginosa FAN1 (PaFAN1) cleaves substrates at ∼3-nt intervals and resolves ICLs. Crystal structures of PaFAN1 bound to various DNA substrates revealed that its conserved basic Arg/Lys patch comprising Arg-228 and Lys-260 recognizes phosphate groups near the 5' terminus of a DNA substrate with a 1-nt flap or a nick. Substitution of Lys-260 did not affect PaFAN1's initial endonuclease activity but significantly decreased its subsequent exonuclease activity and ICL unhooking. The Arg/Lys patch also interacted with phosphates at a 3-nt gap, and this interaction could drive movement of the scissile phosphates into the PaFAN1-active site. In human FAN1, the ICL-resolving activity was not affected by individual disruption of the Arg/Lys patch or basic pocket. However, simultaneous substitution of both FAN1 regions significantly reduced its ICL-resolving activity, suggesting that these two basic regions play a complementary role in ICL repair. On the basis of these findings, we propose a conserved role for two basic regions in FAN1 to guide ICL unhooking and to maintain genomic stability.