Summary information and primary citation
- PDB-id
- 5z9x; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- plant protein-RNA
- Method
- X-ray (2.8 Å)
- Summary
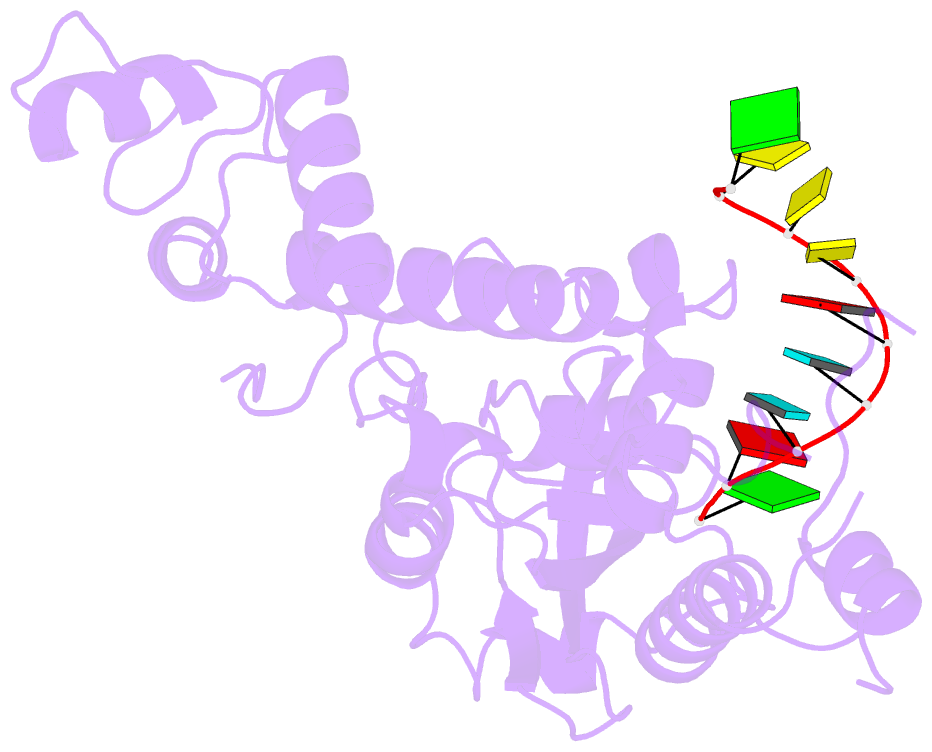
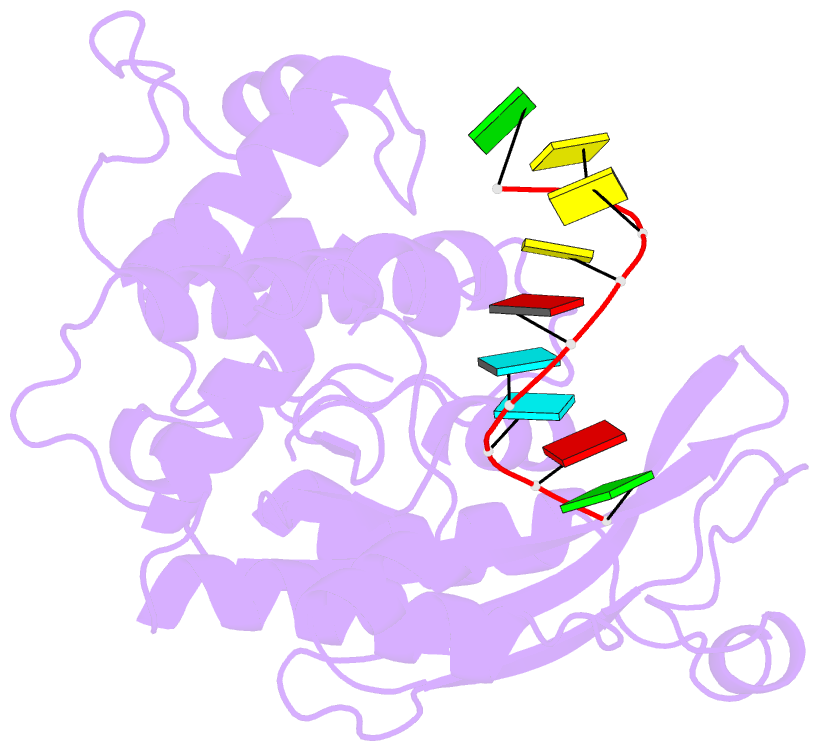
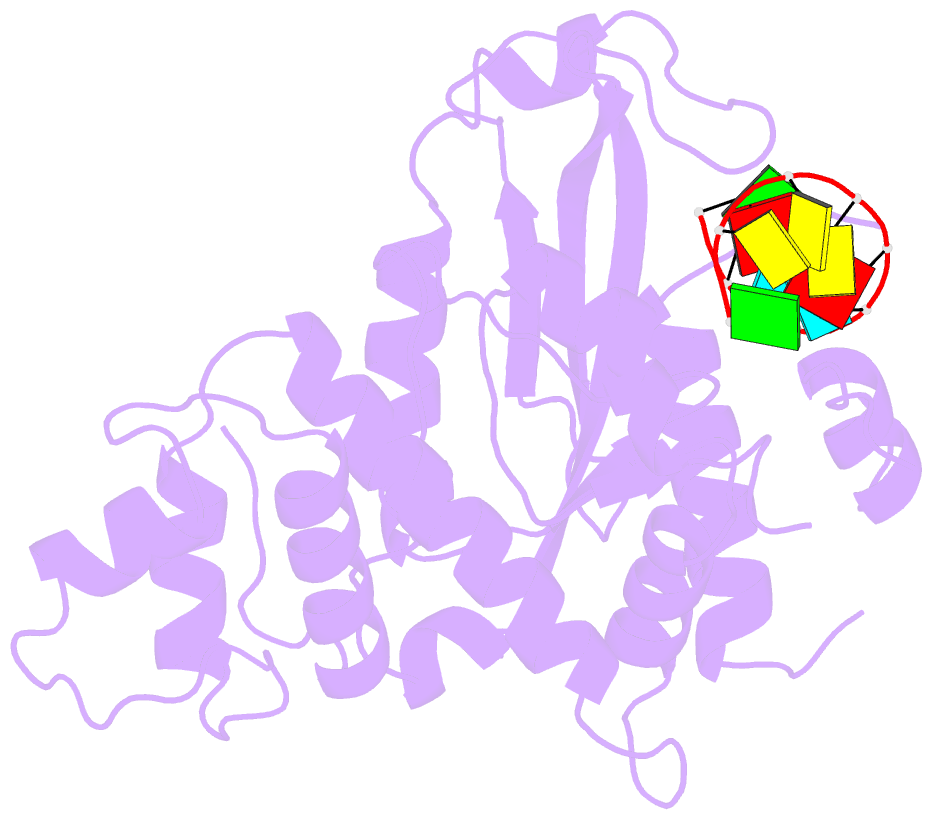
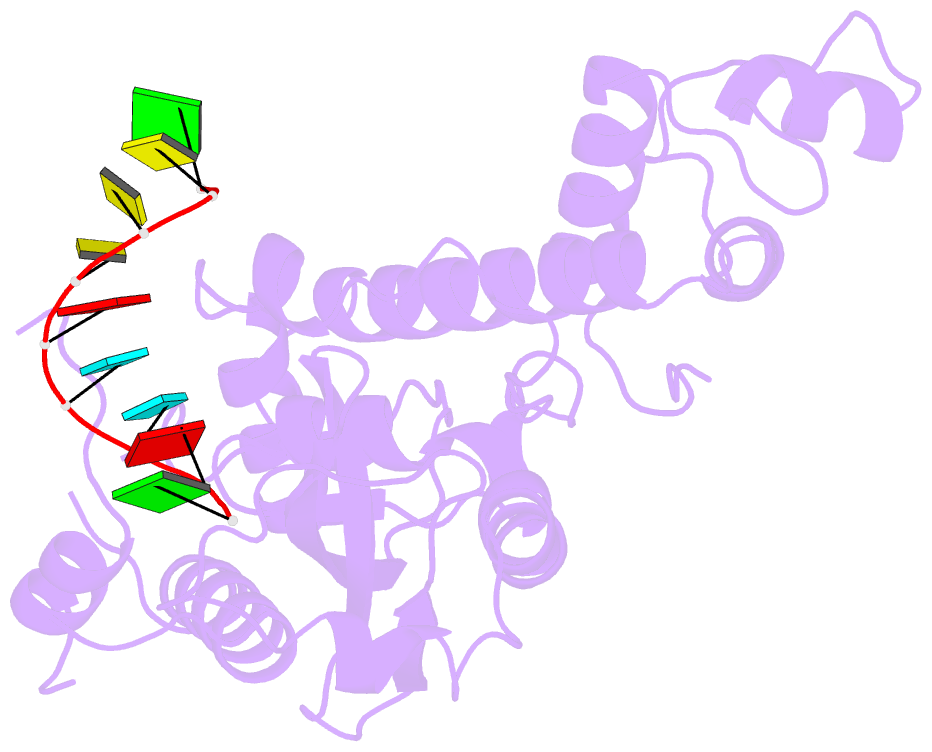
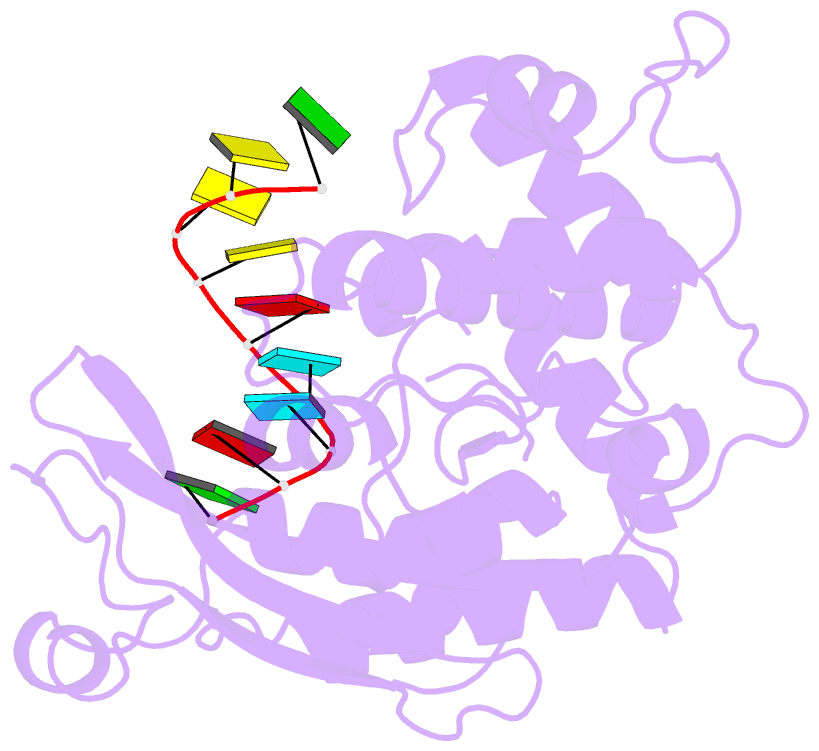
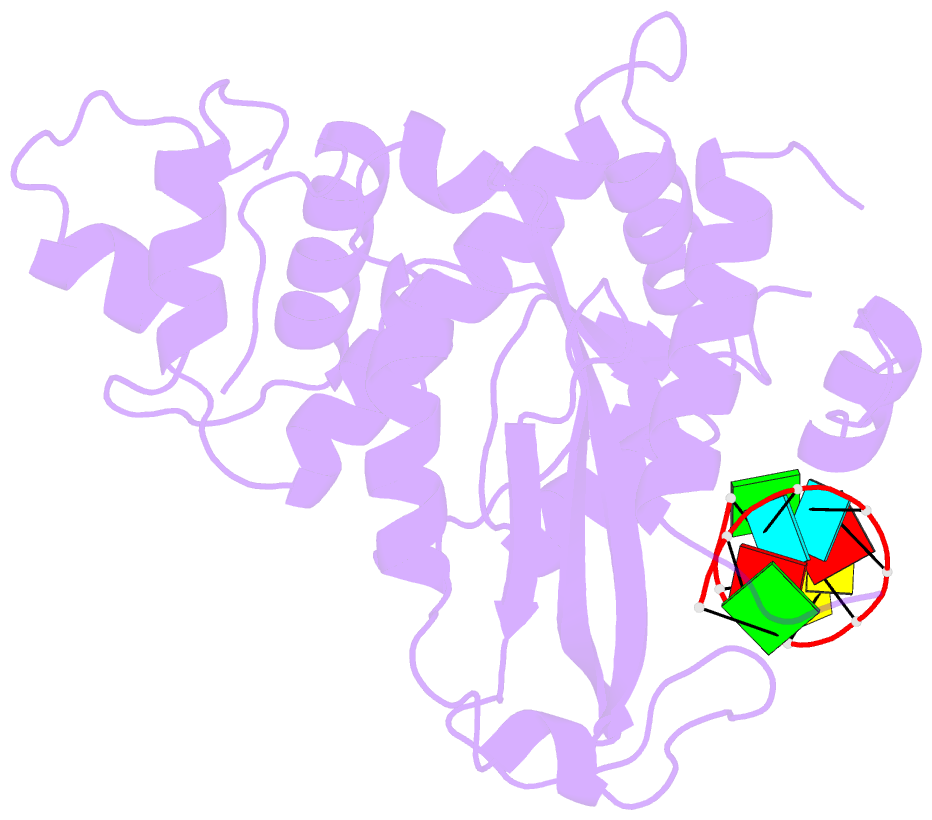
- Arabidopsis small RNA degrading nuclease 1 in complex with an RNA substrate
- Reference
- Chen J, Liu L, You C, Gu J, Ruan W, Zhang L, Gan J, Cao C, Huang Y, Chen X, Ma J (2018): "Structural and biochemical insights into small RNA 3' end trimming by Arabidopsis SDN1." Nat Commun, 9, 3585. doi: 10.1038/s41467-018-05942-7.
- Abstract
- A family of DEDDh 3'→5' exonucleases known as Small RNA Degrading Nucleases (SDNs) initiates the turnover of ARGONAUTE1 (AGO1)-bound microRNAs in Arabidopsis by trimming their 3' ends. Here, we report the crystal structure of Arabidopsis SDN1 (residues 2-300) in complex with a 9 nucleotide single-stranded RNA substrate, revealing that the DEDDh domain forms rigid interactions with the N-terminal domain and binds 4 nucleotides from the 3' end of the RNA via its catalytic pocket. Structural and biochemical results suggest that the SDN1 C-terminal domain adopts an RNA Recognition Motif (RRM) fold and is critical for substrate binding and enzymatic processivity of SDN1. In addition, SDN1 interacts with the AGO1 PAZ domain in an RNA-independent manner in vitro, enabling it to act on AGO1-bound microRNAs. These extensive structural and biochemical studies may shed light on a common 3' end trimming mechanism for 3'→5' exonucleases in the metabolism of small non-coding RNAs.