Summary information and primary citation
- PDB-id
- 5zbx; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.58 Å)
- Summary
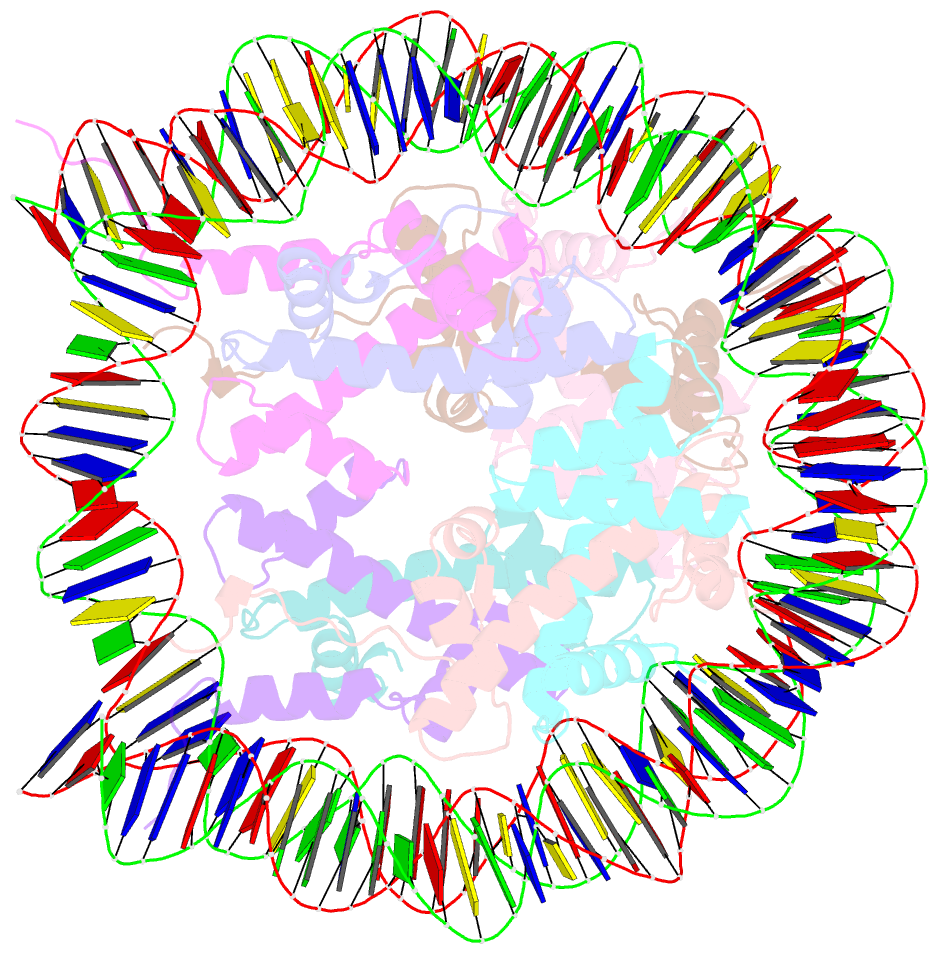
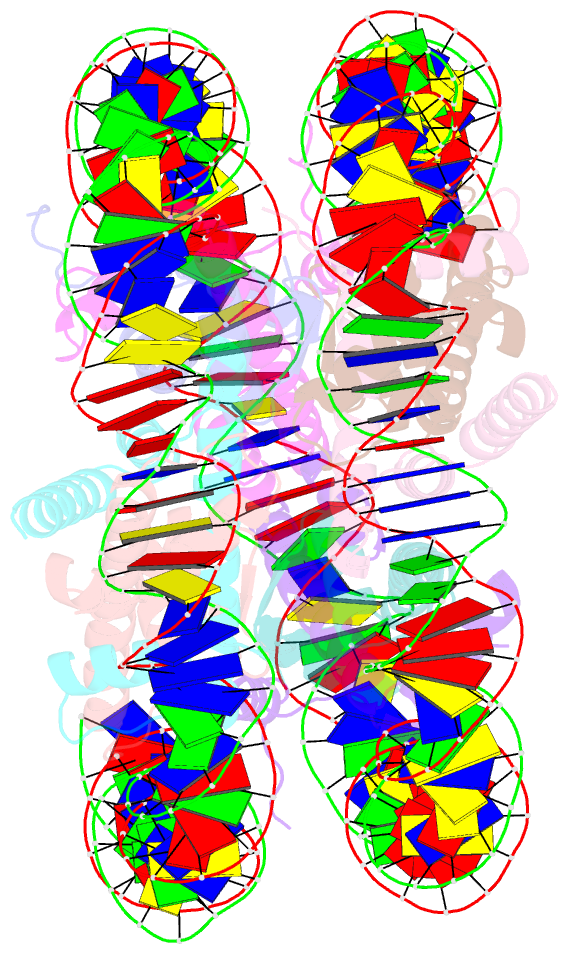
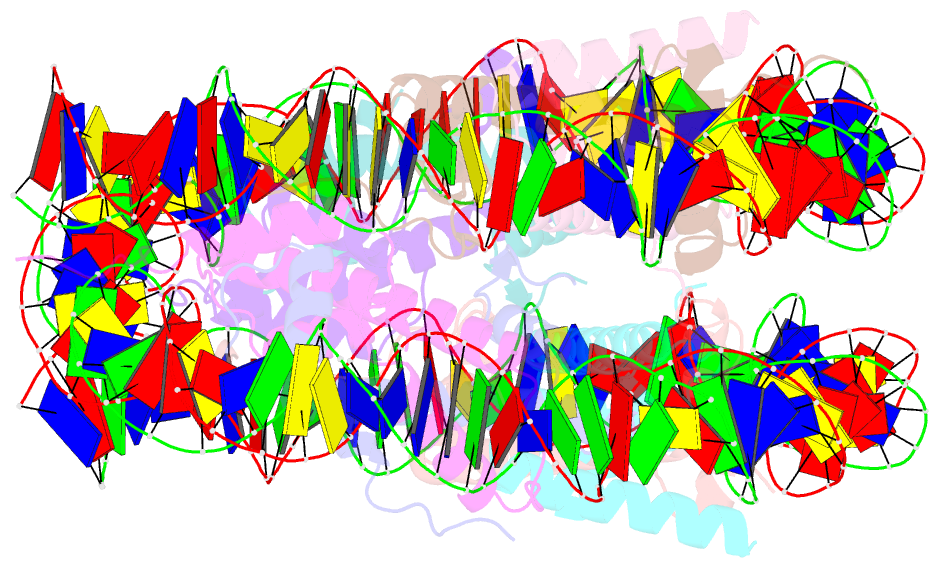
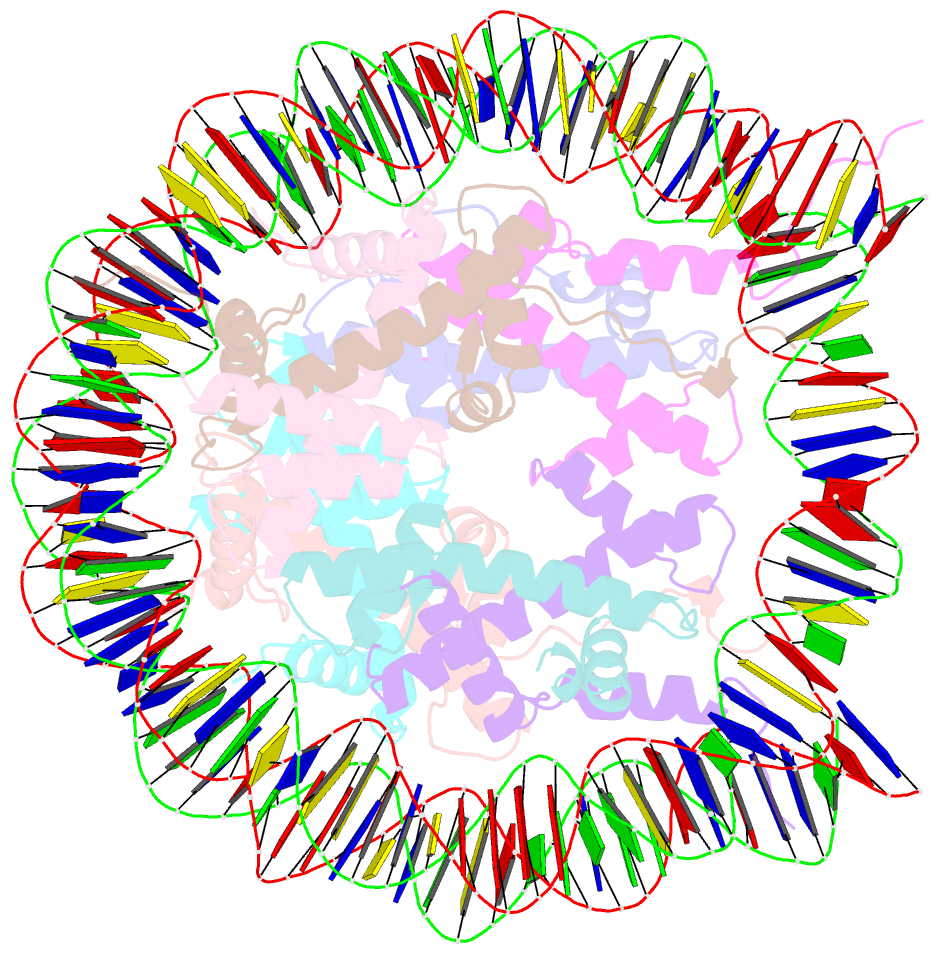
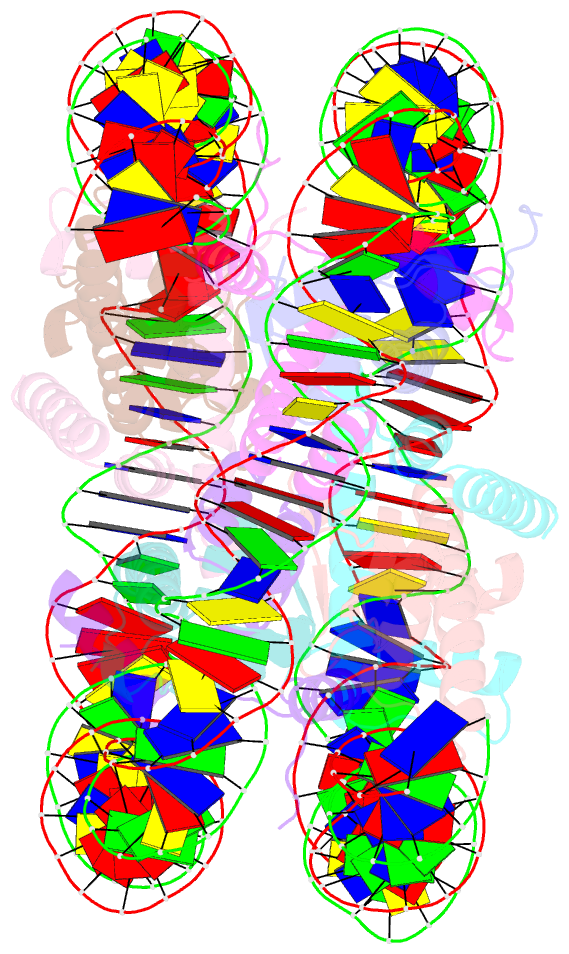
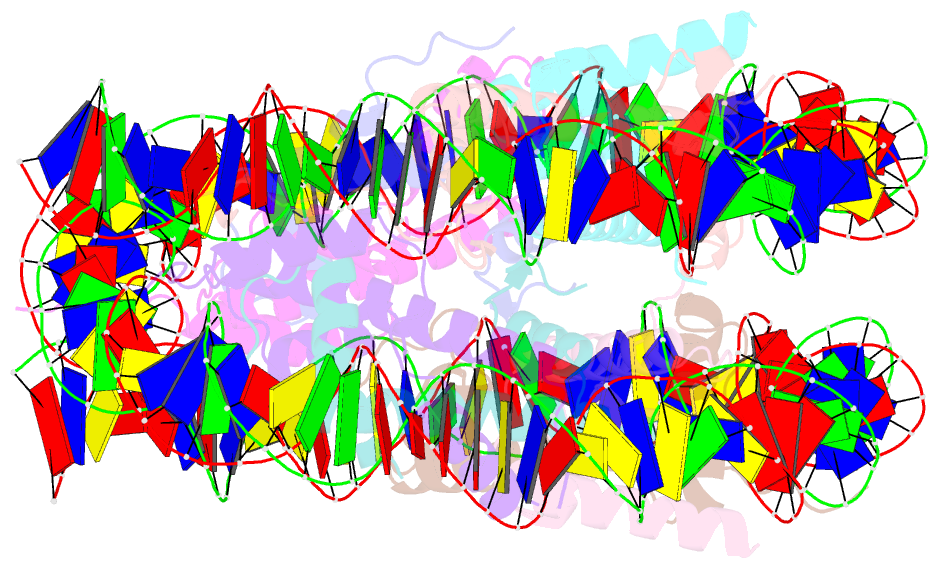
- The crystal structure of the nucleosome containing histone h3.1 catd(v76q, k77d)
- Reference
- Arimura Y, Tachiwana H, Takagi H, Hori T, Kimura H, Fukagawa T, Kurumizaka H (2019): "The CENP-A centromere targeting domain facilitates H4K20 monomethylation in the nucleosome by structural polymorphism." Nat Commun, 10, 576. doi: 10.1038/s41467-019-08314-x.
- Abstract
- Centromeric nucleosomes are composed of the centromere-specific histone H3 variant CENP-A and the core histones H2A, H2B, and H4. To establish a functional kinetochore, histone H4 lysine-20 (H4K20) must be monomethylated, but the underlying mechanism has remained enigmatic. To provide structural insights into H4K20 methylation, we here solve the crystal structure of a nucleosome containing an H3.1-CENP-A chimera, H3.1CATD, which has a CENP-A centromere targeting domain and preserves essential CENP-A functions in vivo. Compared to the canonical H3.1 nucleosome, the H3.1CATD nucleosome exhibits conformational changes in the H4 N-terminal tail leading to a relocation of H4K20. In particular, the H4 N-terminal tail interacts with glutamine-76 and aspartate-77 of canonical H3.1 while these interactions are cancelled in the presence of the CENP-A-specific residues valine-76 and lysine-77. Mutations of valine-76 and lysine-77 impair H4K20 monomethylation both in vitro and in vivo. These findings suggest that a CENP-A-mediated structural polymorphism may explain the preferential H4K20 monomethylation in centromeric nucleosomes.