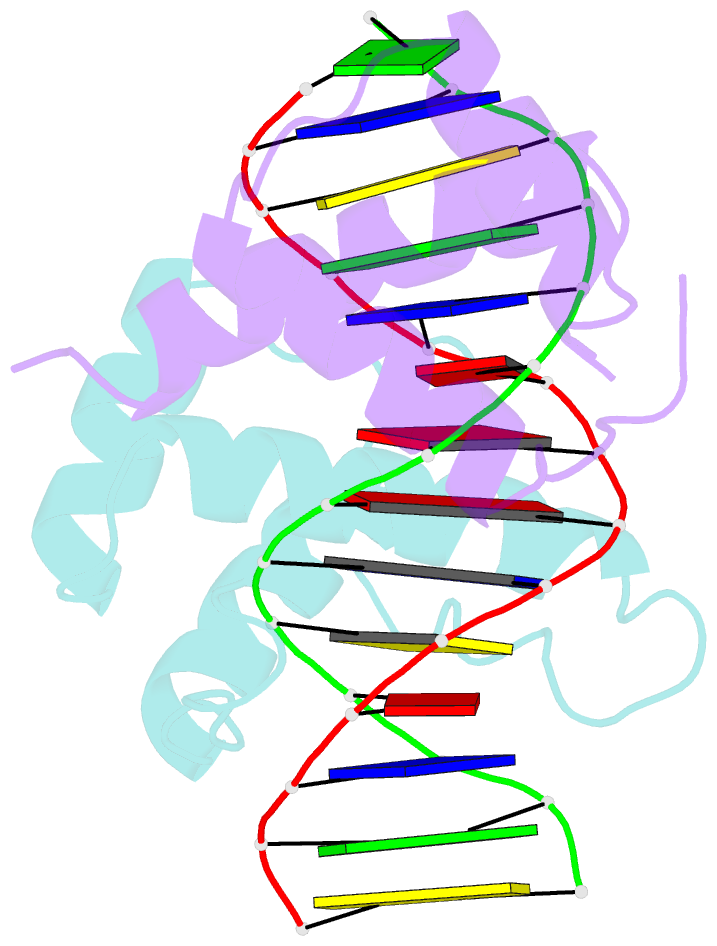
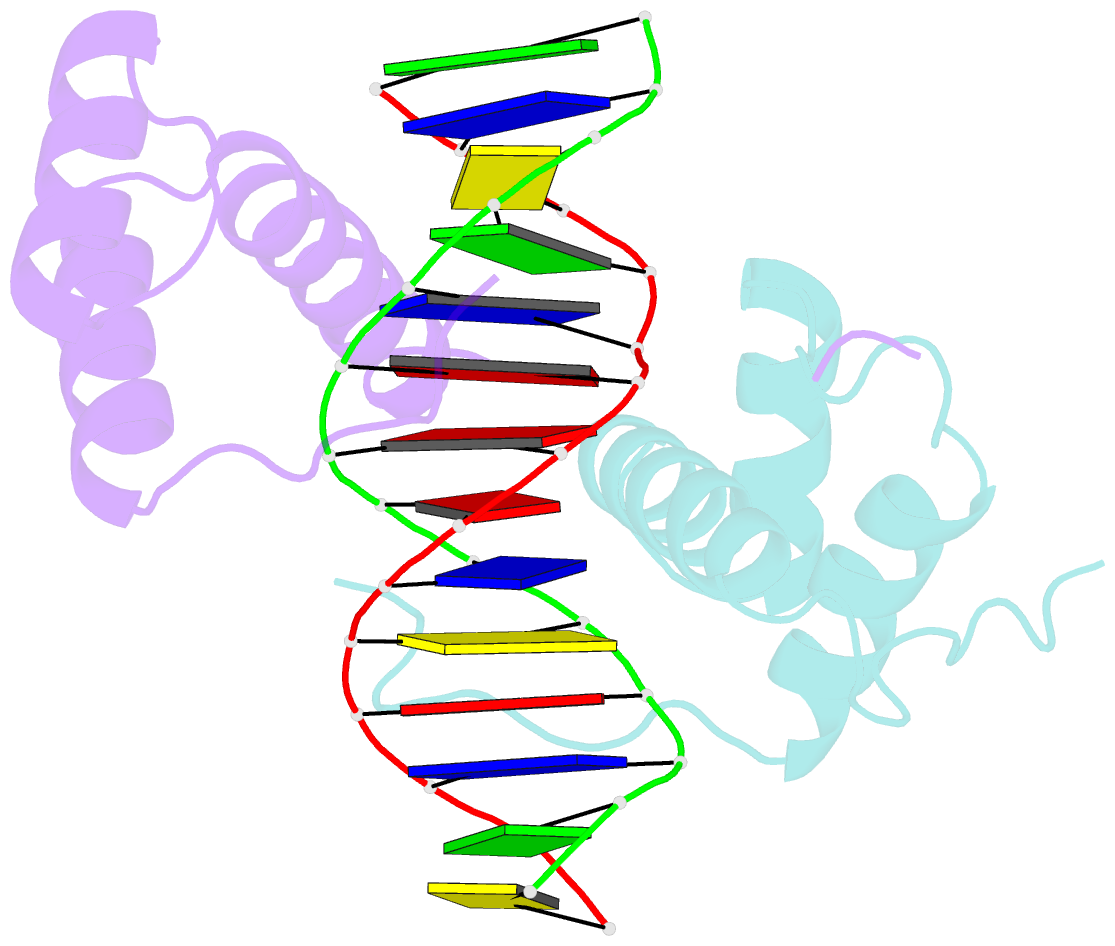
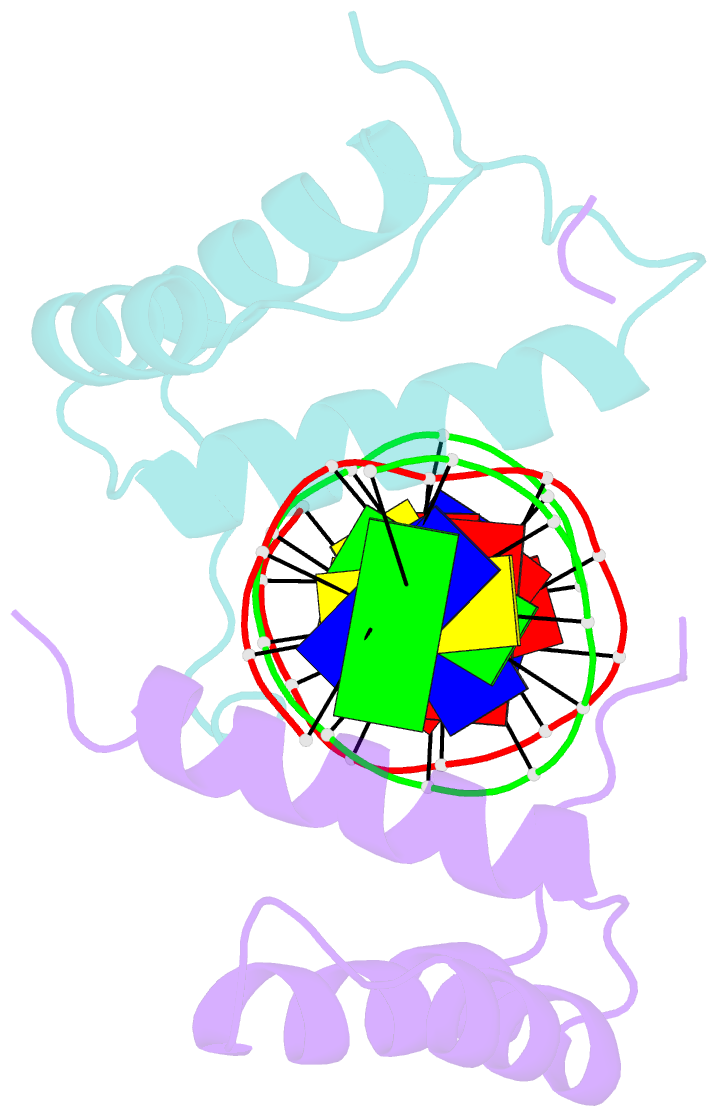
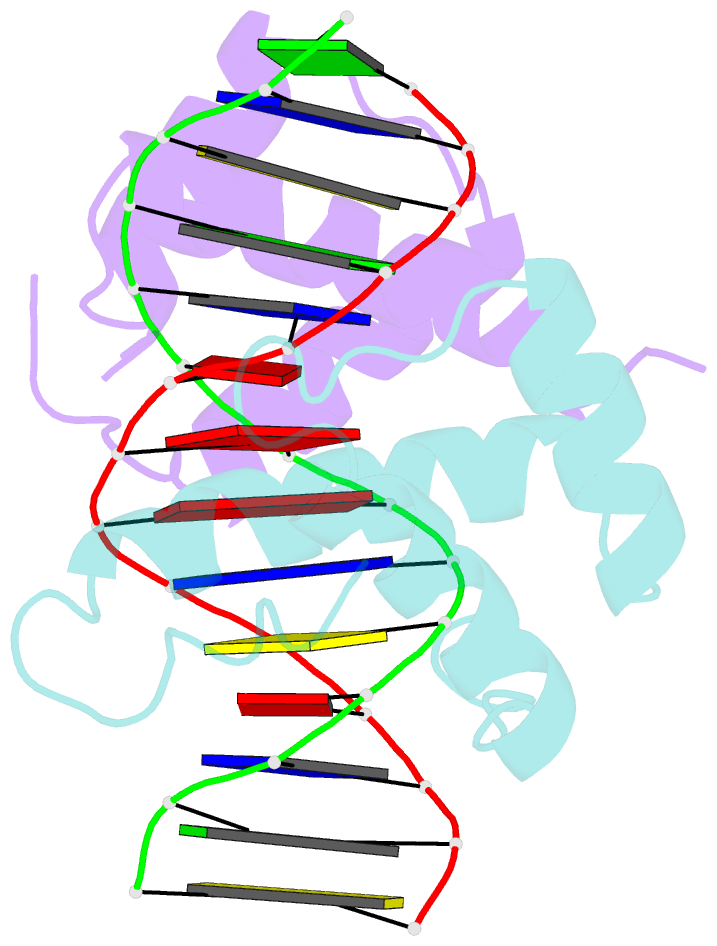
Summary information and primary citation
- PDB-id
- 5zjr; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (3.03 Å)
- Summary
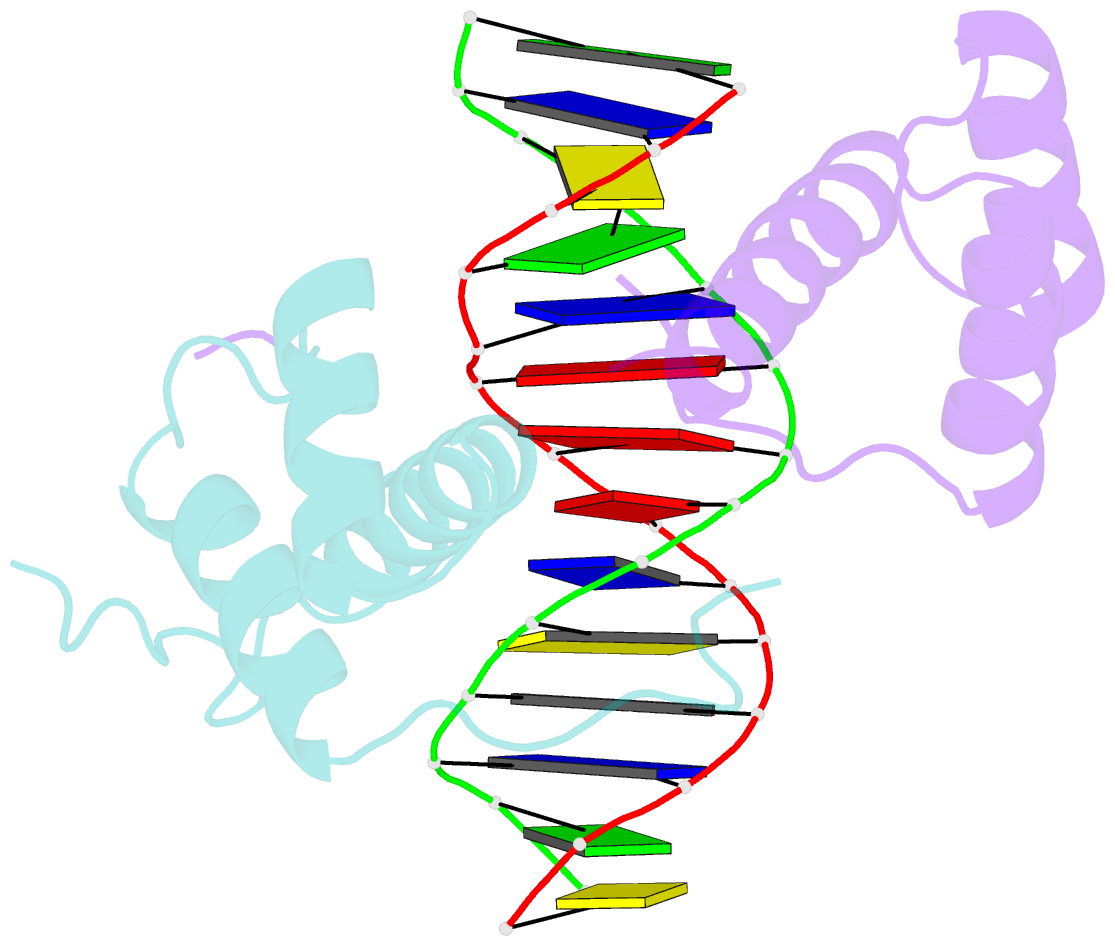
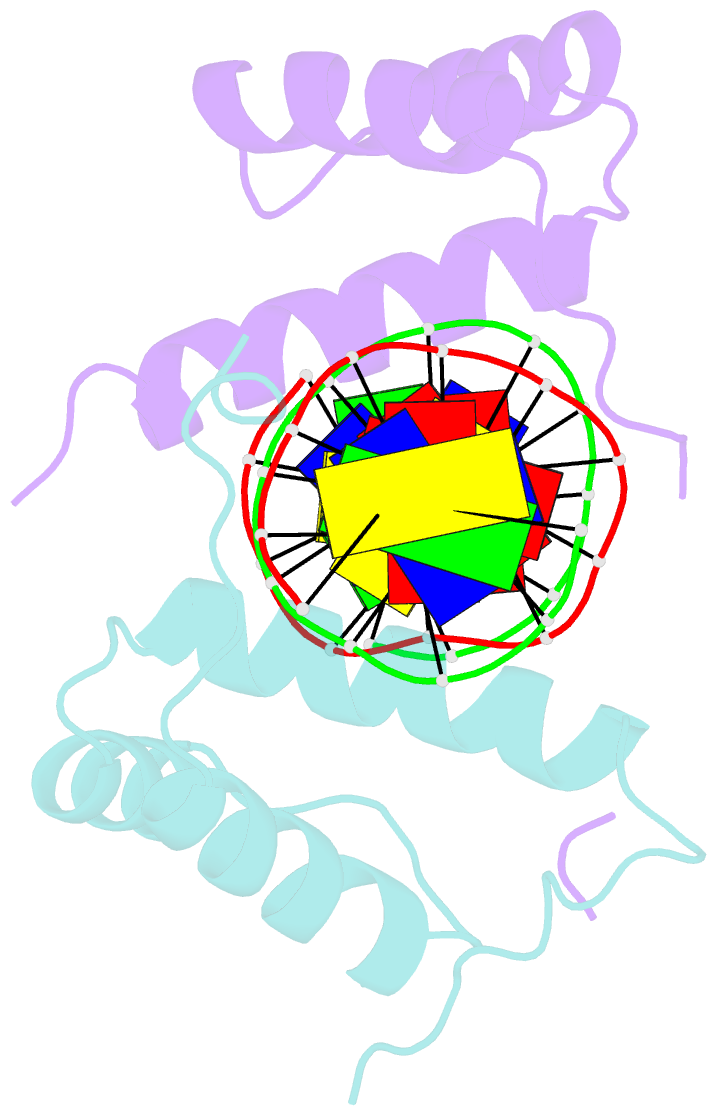
- Structure of abdb-exd complex bound to a 'magenta14' DNA sequence
- Reference
- Zeiske T, Baburajendran N, Kaczynska A, Brasch J, Palmer AG, Shapiro L, Honig B, Mann RS (2018): "Intrinsic DNA Shape Accounts for Affinity Differences between Hox-Cofactor Binding Sites." Cell Rep, 24, 2221-2230. doi: 10.1016/j.celrep.2018.07.100.
- Abstract
- Transcription factors bind to their binding sites over a wide range of affinities, yet how differences in affinity are encoded in DNA sequences is not well understood. Here, we report X-ray crystal structures of four heterodimers of the Hox protein AbdominalB bound with its cofactor Extradenticle to four target DNA molecules that differ in affinity by up to ∼20-fold. Remarkably, despite large differences in affinity, the overall structures are very similar in all four complexes. In contrast, the predicted shapes of the DNA binding sites (i.e., the intrinsic DNA shape) in the absence of bound protein are strikingly different from each other and correlate with affinity: binding sites that must change conformations upon protein binding have lower affinities than binding sites that have more optimal conformations prior to binding. Together, these observations suggest that intrinsic differences in DNA shape provide a robust mechanism for modulating affinity without affecting other protein-DNA interactions.