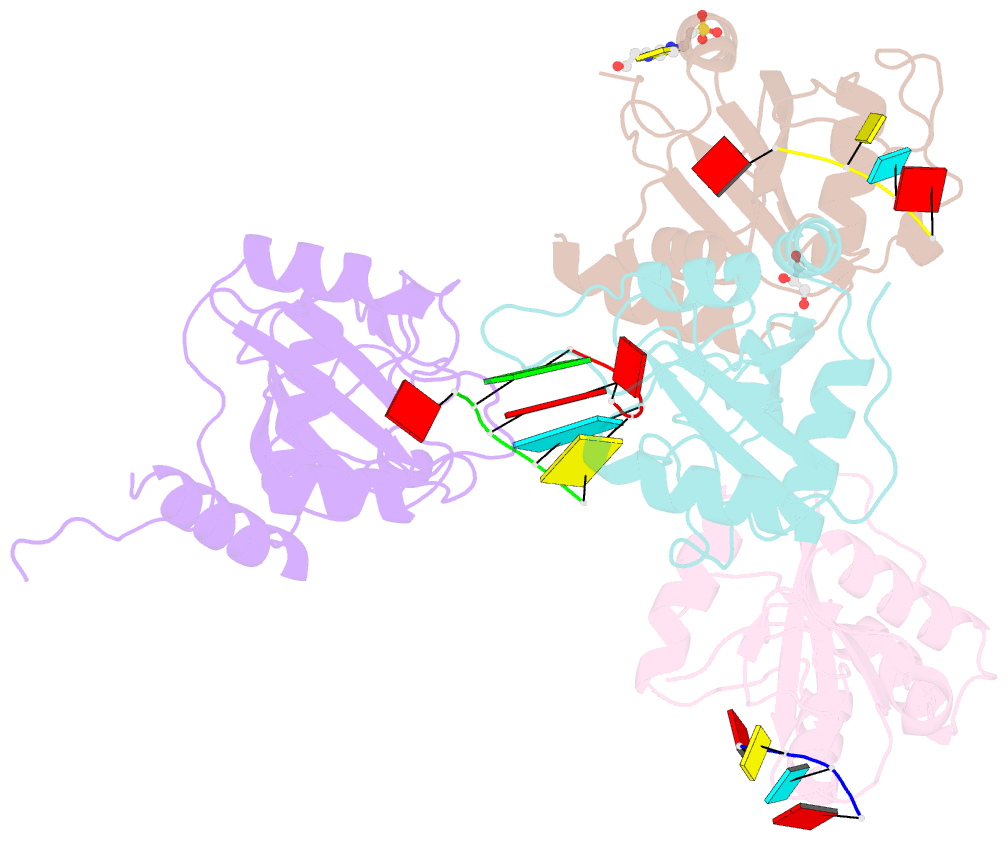
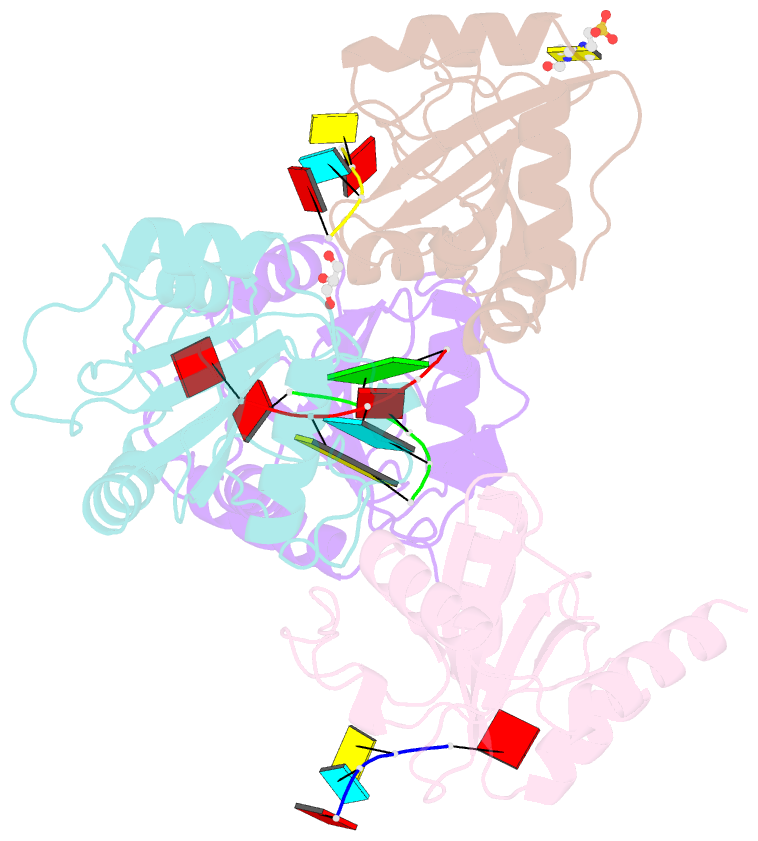
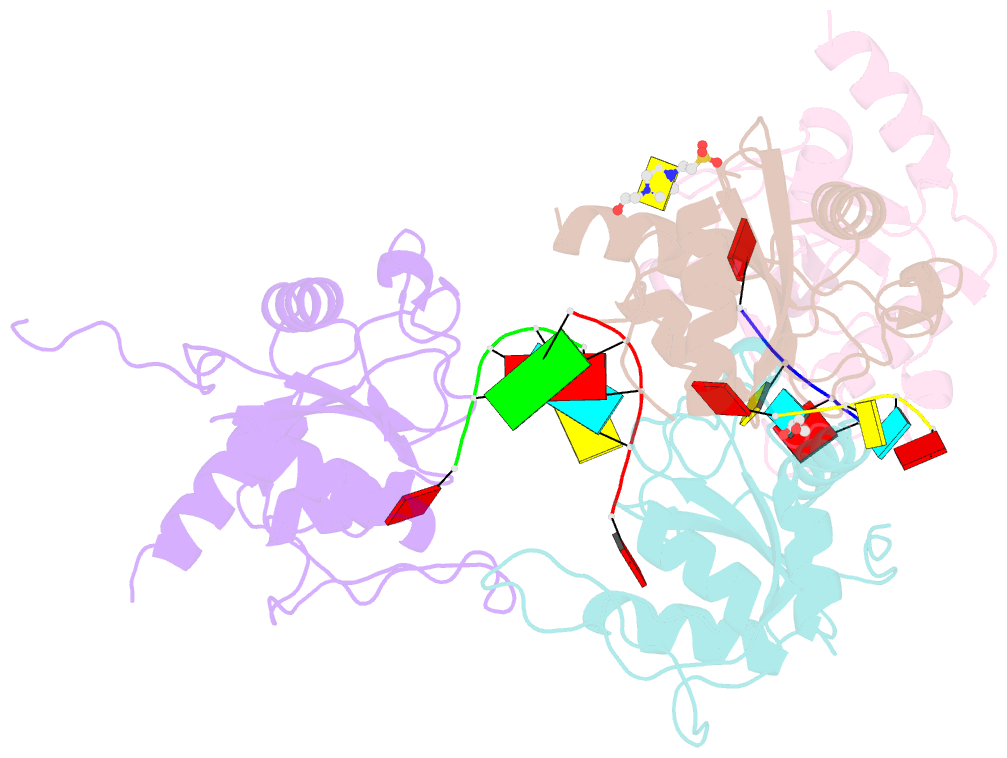
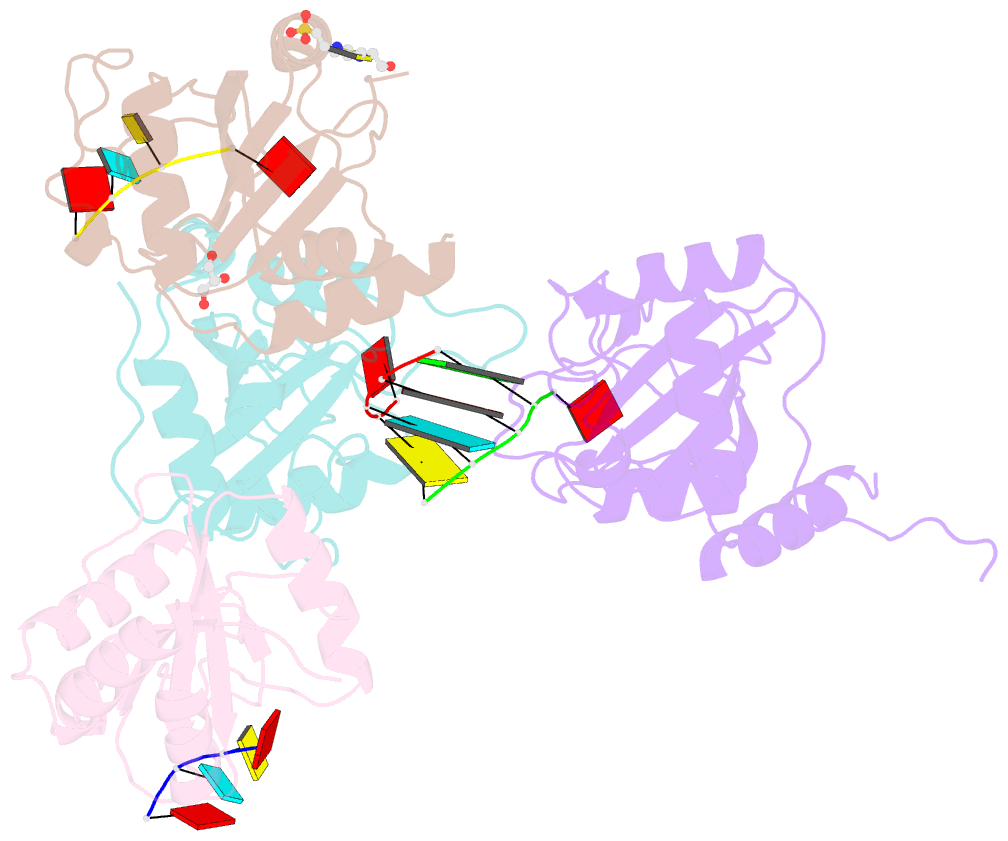
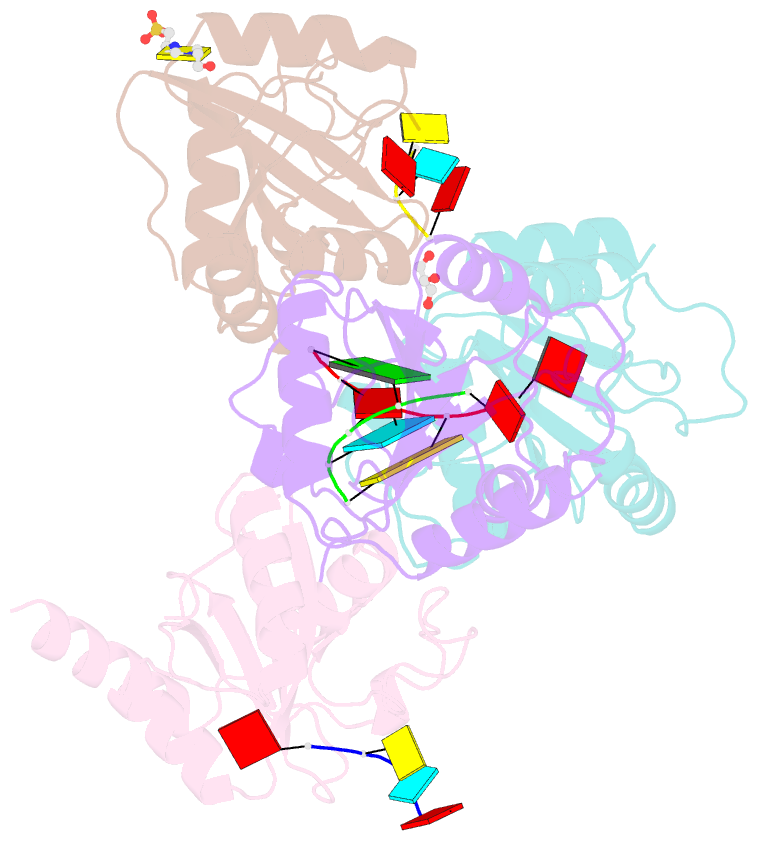
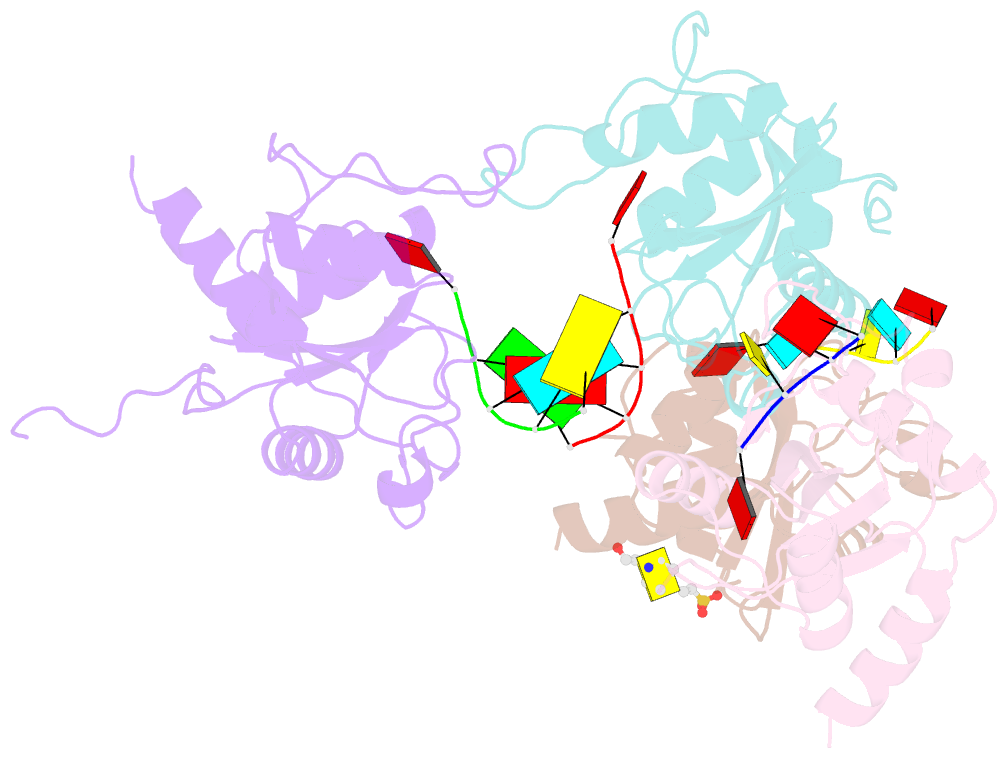
Summary information and primary citation
- PDB-id
- 5zuu; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- X-ray (1.95 Å)
- Summary
- Crystal structure of atcpsf30 yth domain in complex with 10mer m6a-modified RNA
- Reference
- Hou Y, Sun J, Wu B, Gao Y, Nie H, Nie Z, Quan S, Wang Y, Cao X, Li S (2021): "CPSF30-L-mediated recognition of mRNA m6A modification controls alternative polyadenylation of nitrate signaling-related gene transcripts in Arabidopsis." Mol Plant. doi: 10.1016/j.molp.2021.01.013.
- Abstract
- N6-methyladenosine (m6A), a ubiquitous internal modification of eukaryotic mRNAs, plays a vital role in almost every aspect of mRNA metabolism. However, there is little evidence documenting the role of m6A in regulating alternative polyadenylation (APA) in plants. APA is controlled by a large protein-RNA complex with many components, including CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR30 (CPSF30). In Arabidopsis, CPSF30 has two isoforms and the longer isoform (CPSF30-L) contains a YT512-B Homology (YTH) domain, which is unique to plants. In this study, we showed that CPSF30-L YTH domain binds to m6A in vitro. In the cpsf30-2 mutant, the transcripts of many genes including several important nitrate signaling-related genes had shifts in polyadenylation sites that were correlated with m6A peaks, indicating that these gene transcripts carrying m6A tend to be regulated by APA. Wild-type CPSF30-L could rescue the defects in APA and nitrate metabolism in cpsf30-2, but m6A-binding-defective mutants of CPSF30-L could not. Taken together, our results demonstrated that m6A modification regulates APA in Arabidopsis and revealed that the m6A reader CPSF30-L affects nitrate signaling by controlling APA, shedding new light on the roles of the m6A modification during RNA 3'-end processing in nitrate metabolism.