Summary information and primary citation
- PDB-id
- 5zwo; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- splicing
- Method
- cryo-EM (3.9 Å)
- Summary
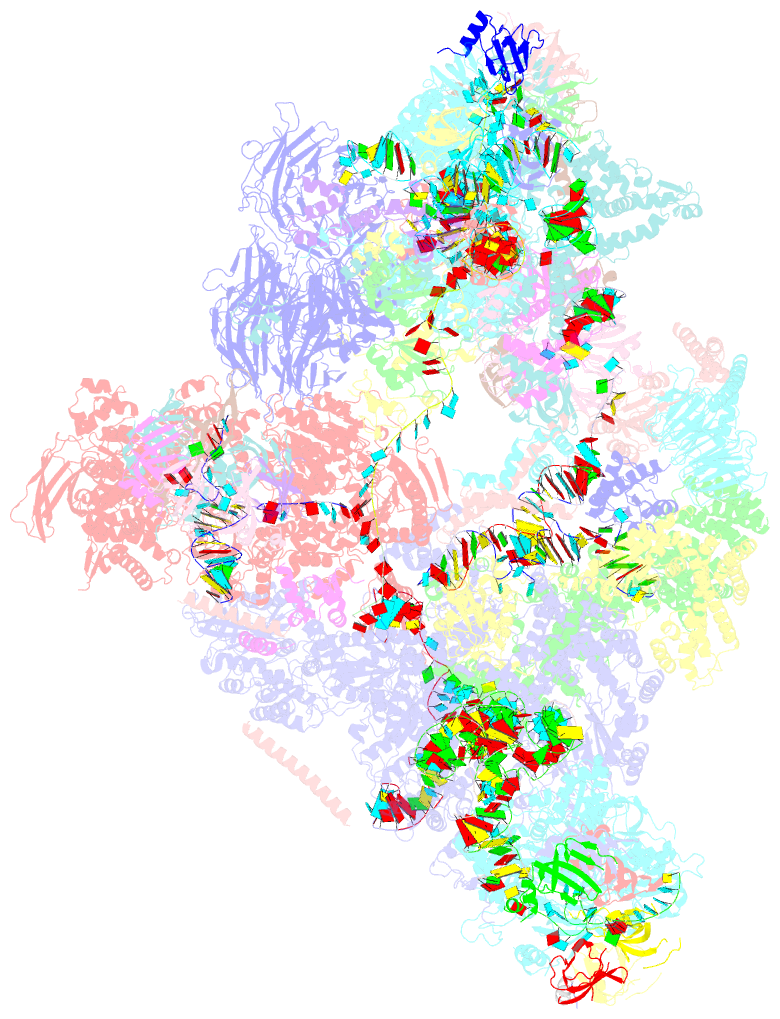
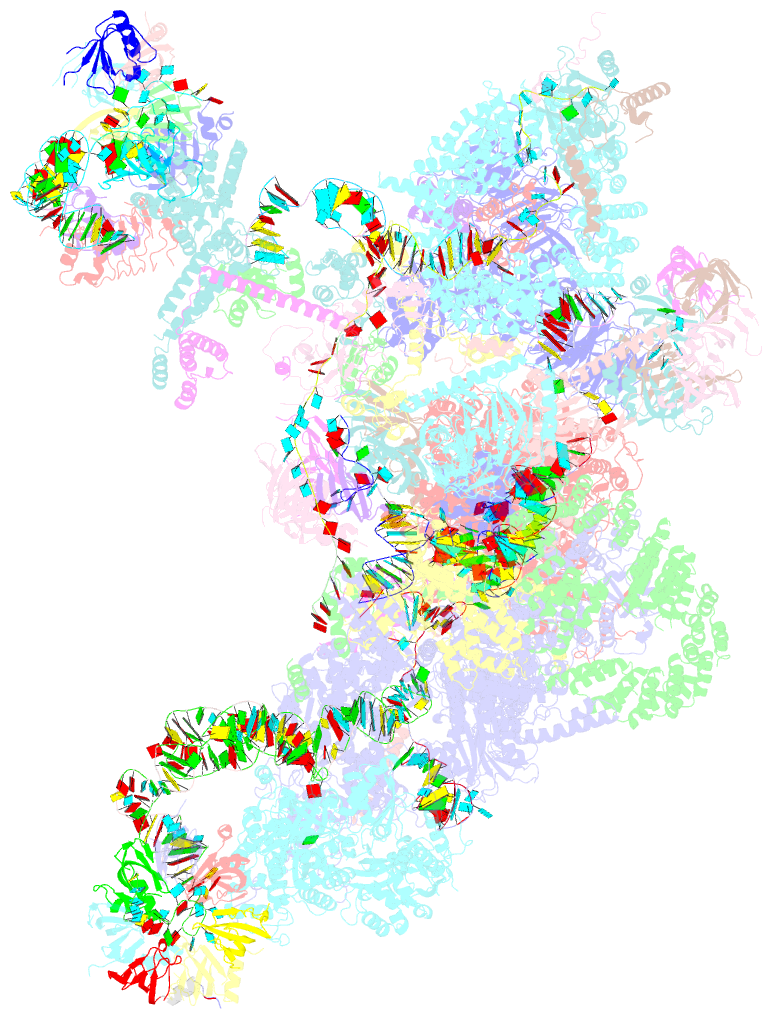
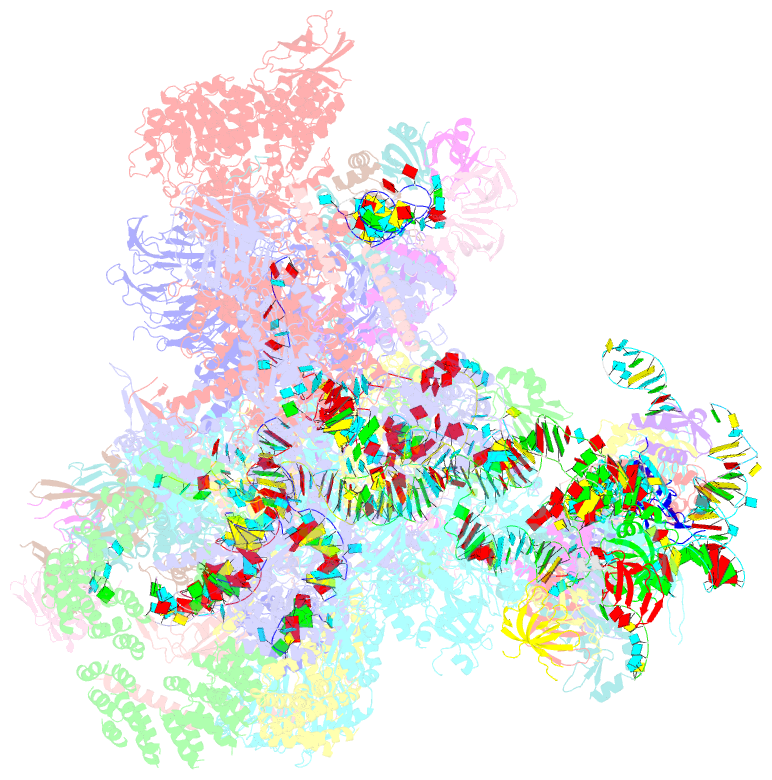
- cryo-EM structure of the yeast b complex at average resolution of 3.9 angstrom
- Reference
- Bai R, Wan R, Yan C, Lei J, Shi Y (2018): "Structures of the fully assembledSaccharomyces cerevisiaespliceosome before activation." Science, 360, 1423-1429. doi: 10.1126/science.aau0325.
- Abstract
- The precatalytic spliceosome (B complex) is preceded by the pre-B complex. Here we report the cryo-electron microscopy structures of the Saccharomyces cerevisiae pre-B and B complexes at average resolutions of 3.3 to 4.6 and 3.9 angstroms, respectively. In the pre-B complex, the duplex between the 5' splice site (5'SS) and U1 small nuclear RNA (snRNA) is recognized by Yhc1, Luc7, and the Sm ring. In the B complex, U1 small nuclear ribonucleoprotein is dissociated, the 5'-exon-5'SS sequences are translocated near U6 snRNA, and three B-specific proteins may orient the precursor messenger RNA. In both complexes, U6 snRNA is anchored to loop I of U5 snRNA, and the duplex between the branch point sequence and U2 snRNA is recognized by the SF3b complex. Structural analysis reveals the mechanism of assembly and activation for the yeast spliceosome.