Summary information and primary citation
- PDB-id
- 6a4b; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- X-ray (2.7 Å)
- Summary
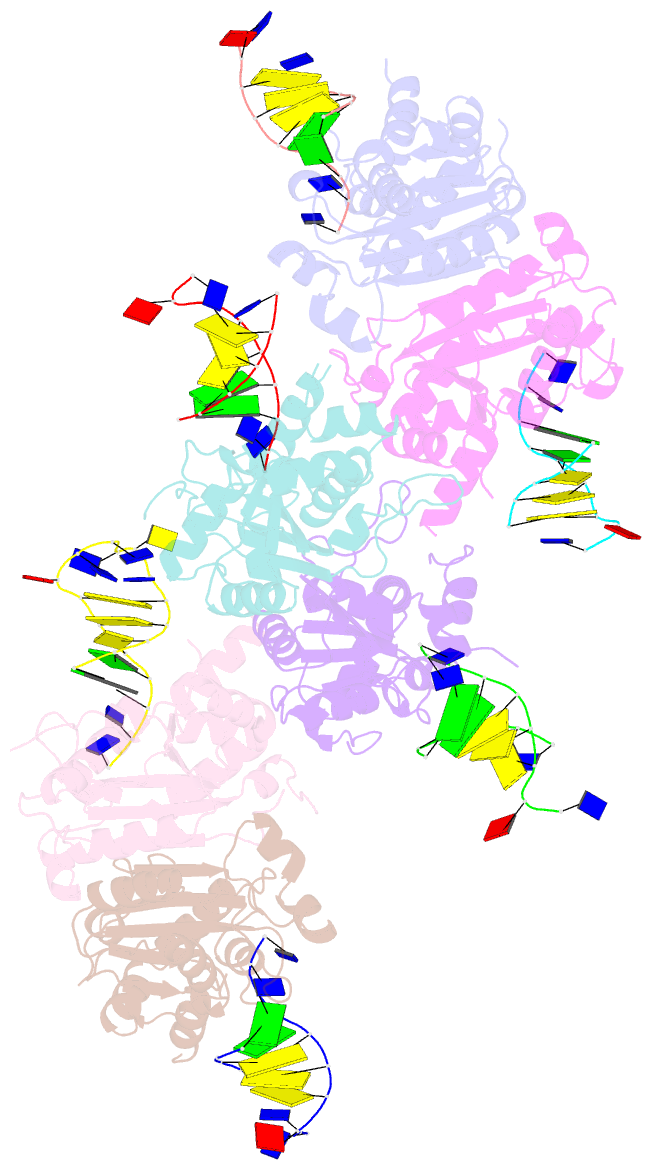
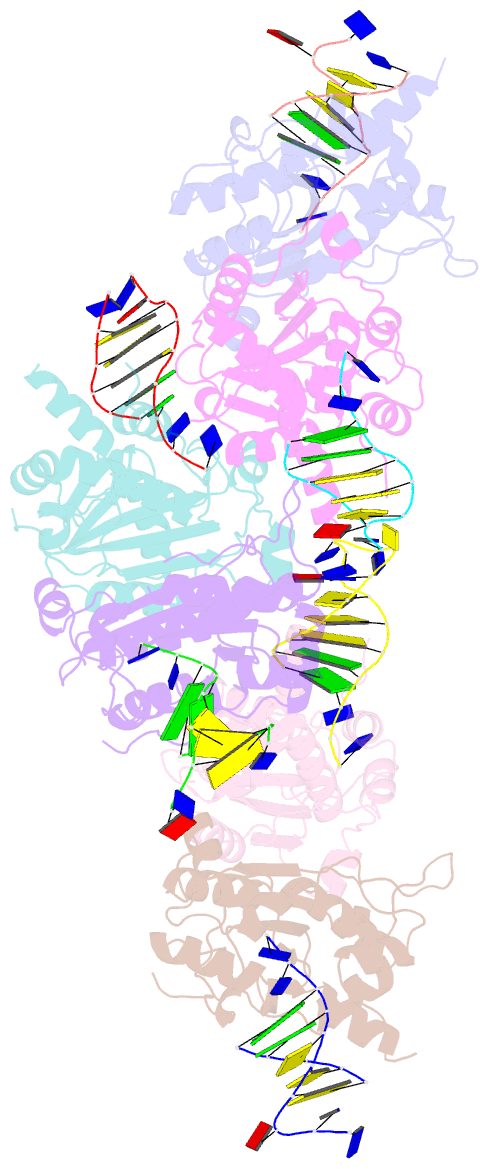
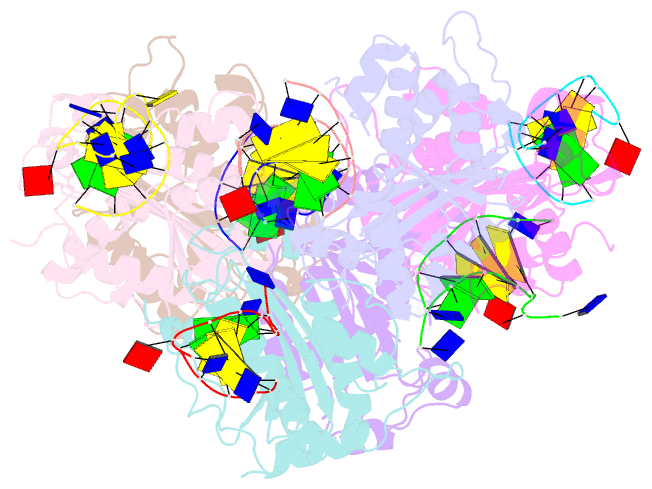
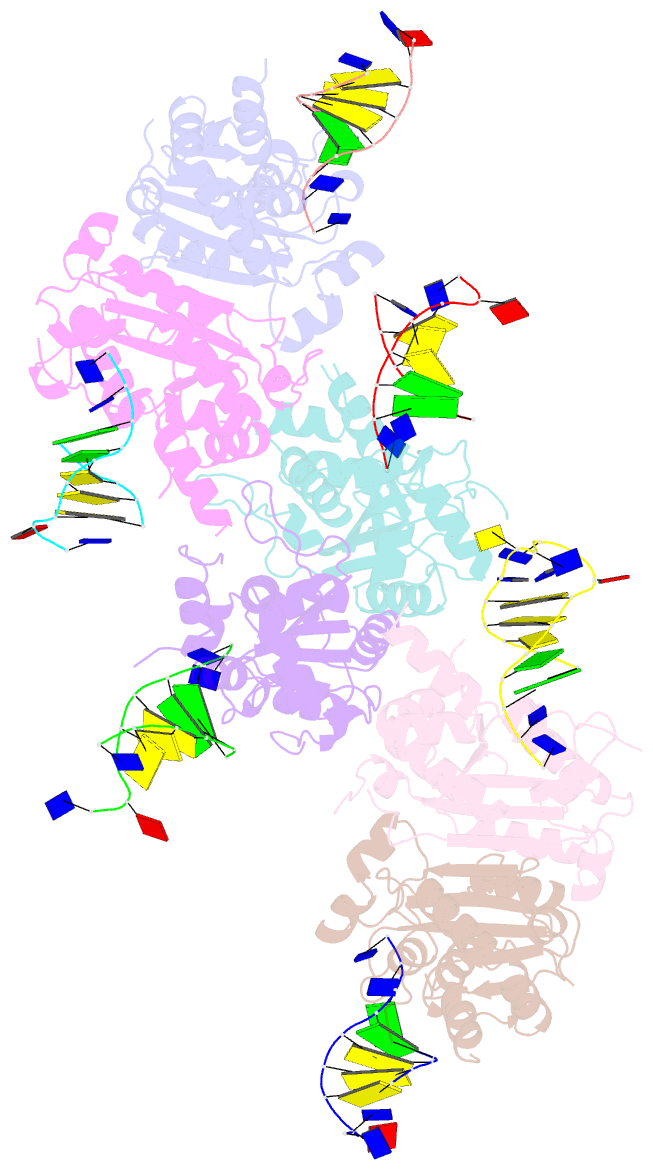
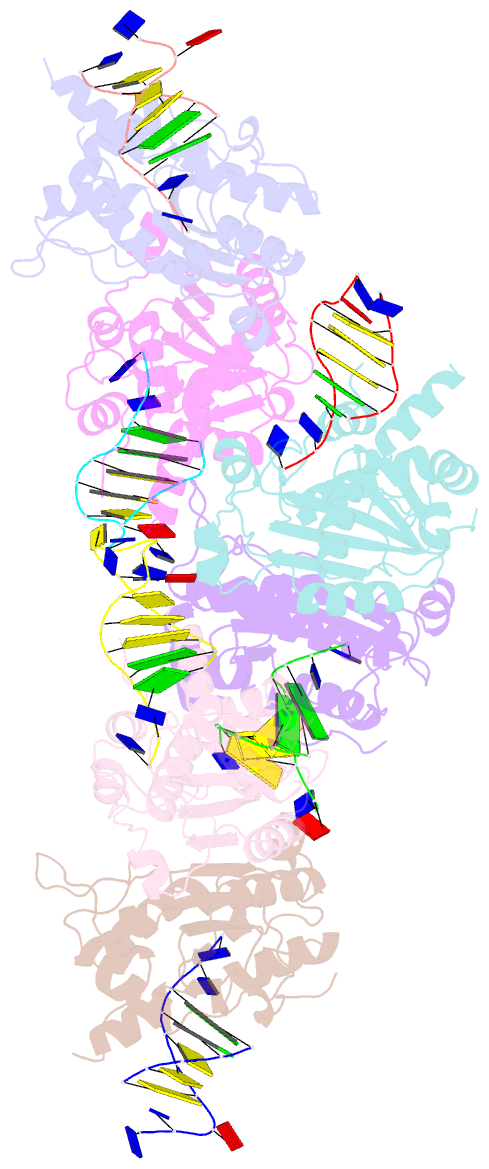
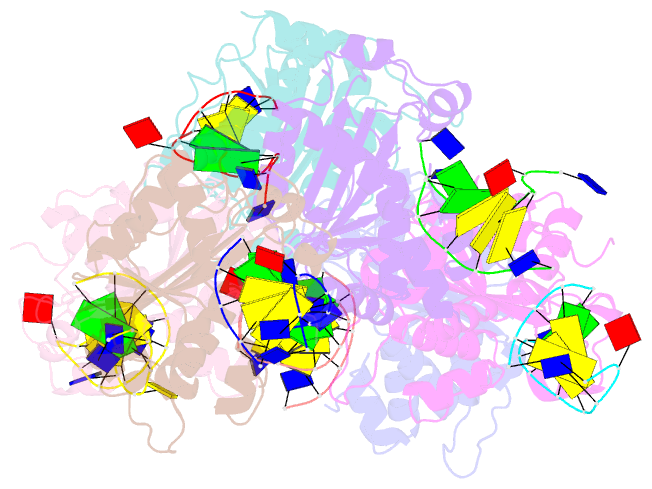
- Structure of trex2 in complex with a duplex DNA with 2 nucleotide 3'-overhang
- Reference
- Cheng HL, Lin CT, Huang KW, Wang S, Lin YT, Toh SI, Hsiao YY (2018): "Structural insights into the duplex DNA processing of TREX2." Nucleic Acids Res., 46, 12166-12176. doi: 10.1093/nar/gky970.
- Abstract
- The three prime repair exonuclease 2 (TREX2) is an essential 3'-to-5' exonuclease that functions in cell proliferation, genome integrity and skin homeostasis maintenance. The abnormal expression level of TREX2 can result in broken chromosome, increased susceptibility to skin carcinogenesis and Psoriasis. However, the molecular mechanisms of how TREX2 binds and processes its natural substrates, dsDNA or chromosomal DNA, to maintain genome stability remain unclear. In this study, we present four new crystal structures: apo-TREX2, TREX2 in complex with two different dsDNA substrates, and TREX2 in complex with a processed dsDNA product. Analysis of the structures reveals that TREX2 stacks with the 5'-terminal of dsDNA by a Leu20-Pro21-Asn22 cluster for precisely trimming the 3'-overhang. In addition, TREX2 specifically interacts with the non-scissile strand of dsDNA by an α-helix-loop region. The unique interaction patterns of the TREX2-dsDNA complex highlight the requirement of long double-stranded region for TREX2 binding and provide evidence of the functional role of TREX2 in processing chromosomal DNA. Moreover, the non-processive property of TREX2 is elucidated by the structure of TREX2-product complex. Our work discloses the first structural basis of the molecular interactions between TREX2 and its substrates and unravels the mechanistic actions of TREX2.