Summary information and primary citation
- PDB-id
- 6a4e; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase
- Method
- X-ray (2.45 Å)
- Summary
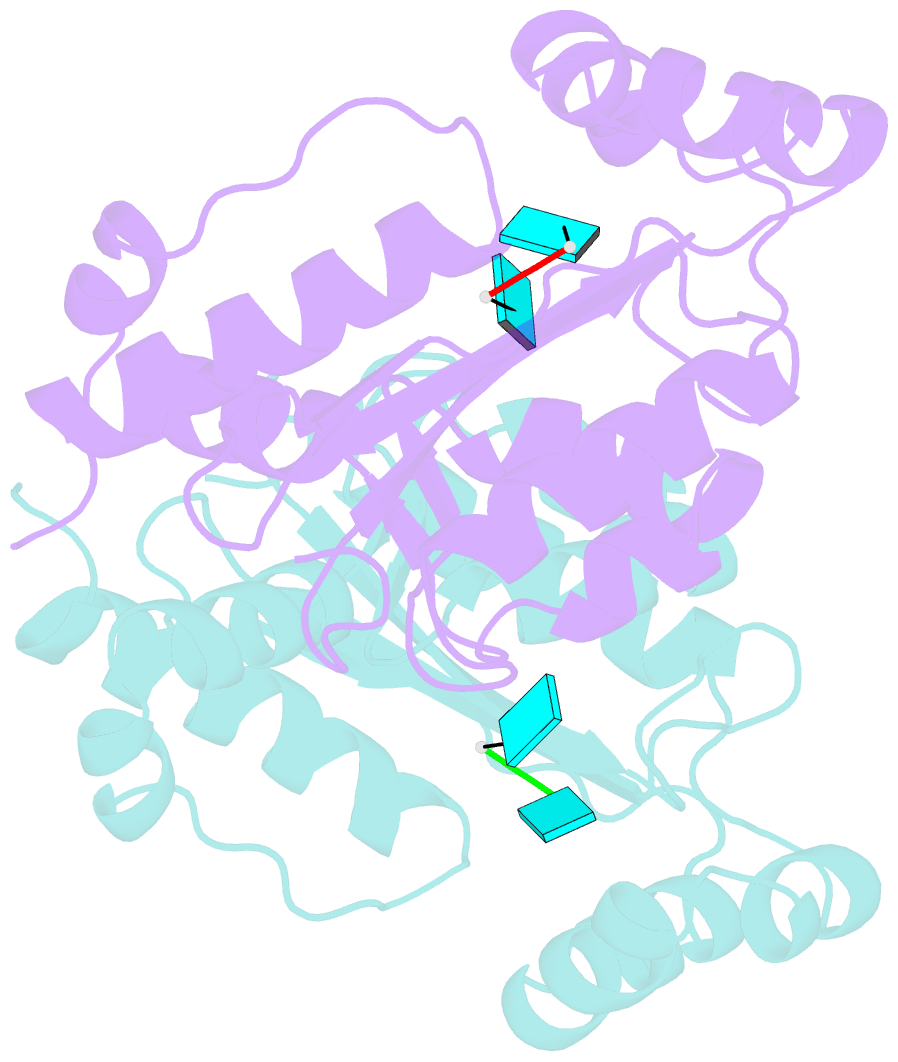
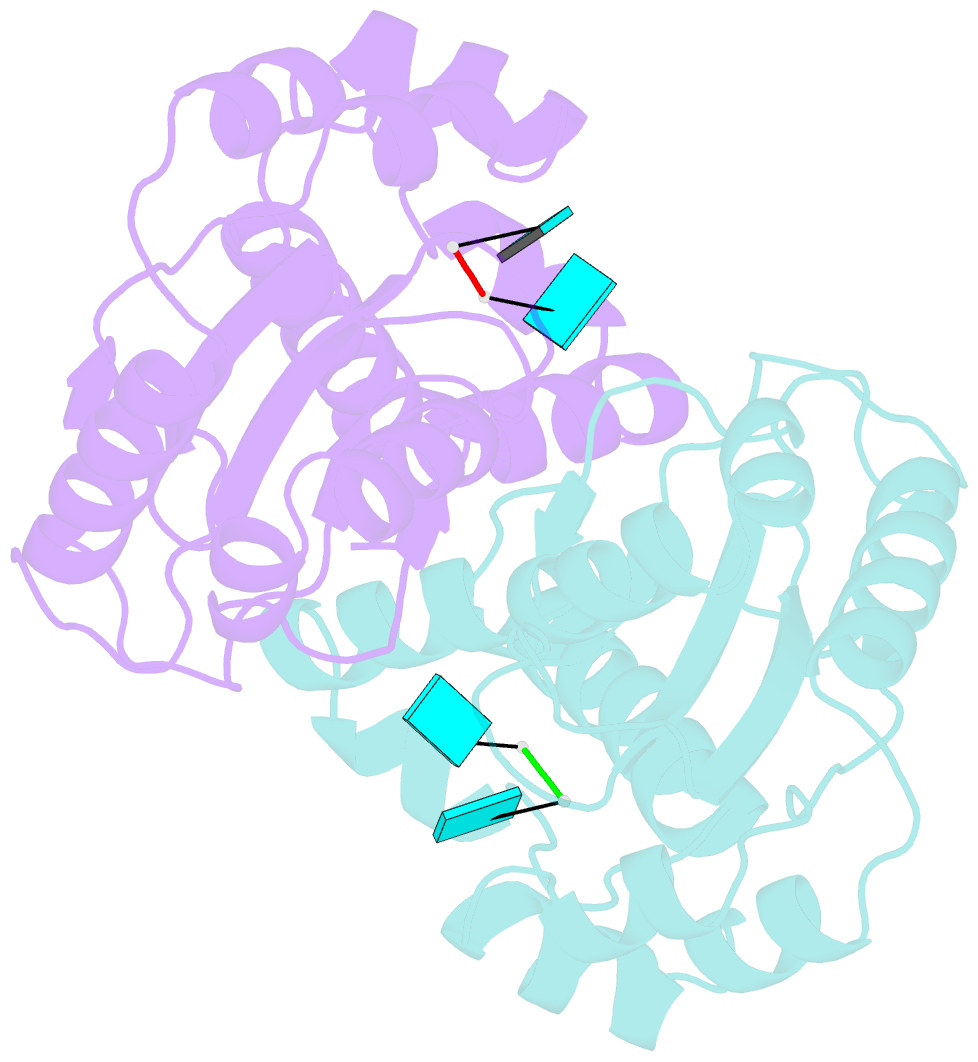
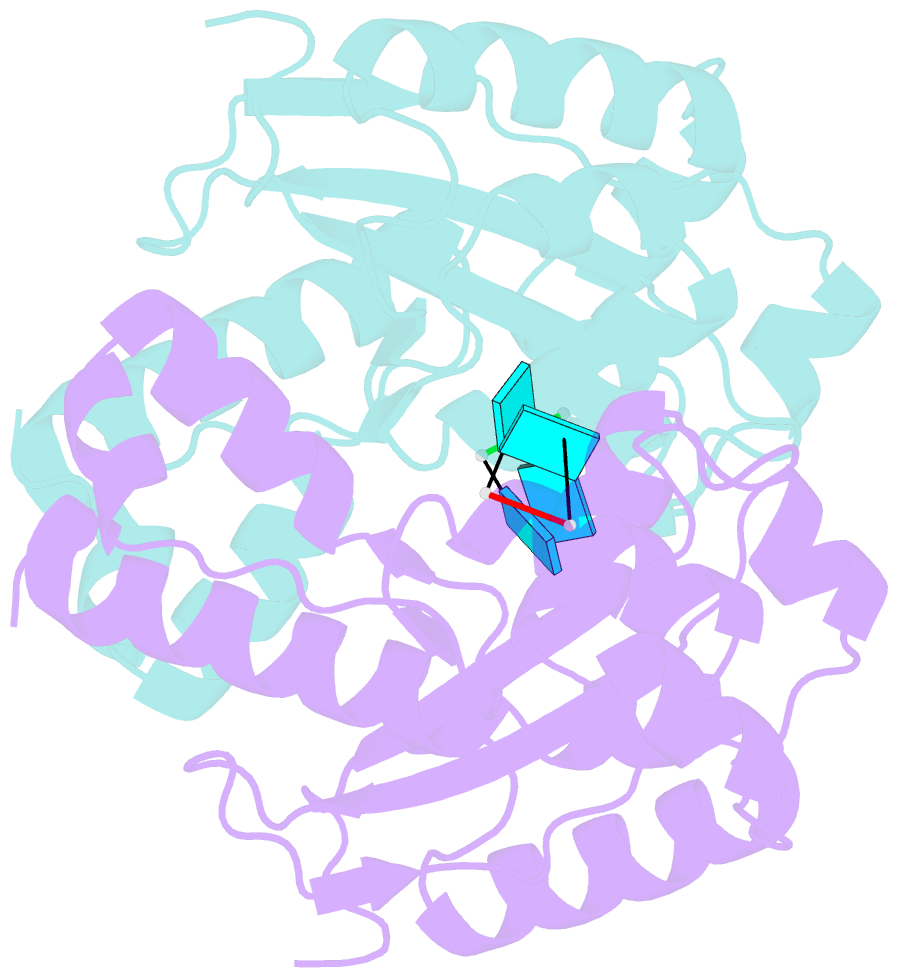
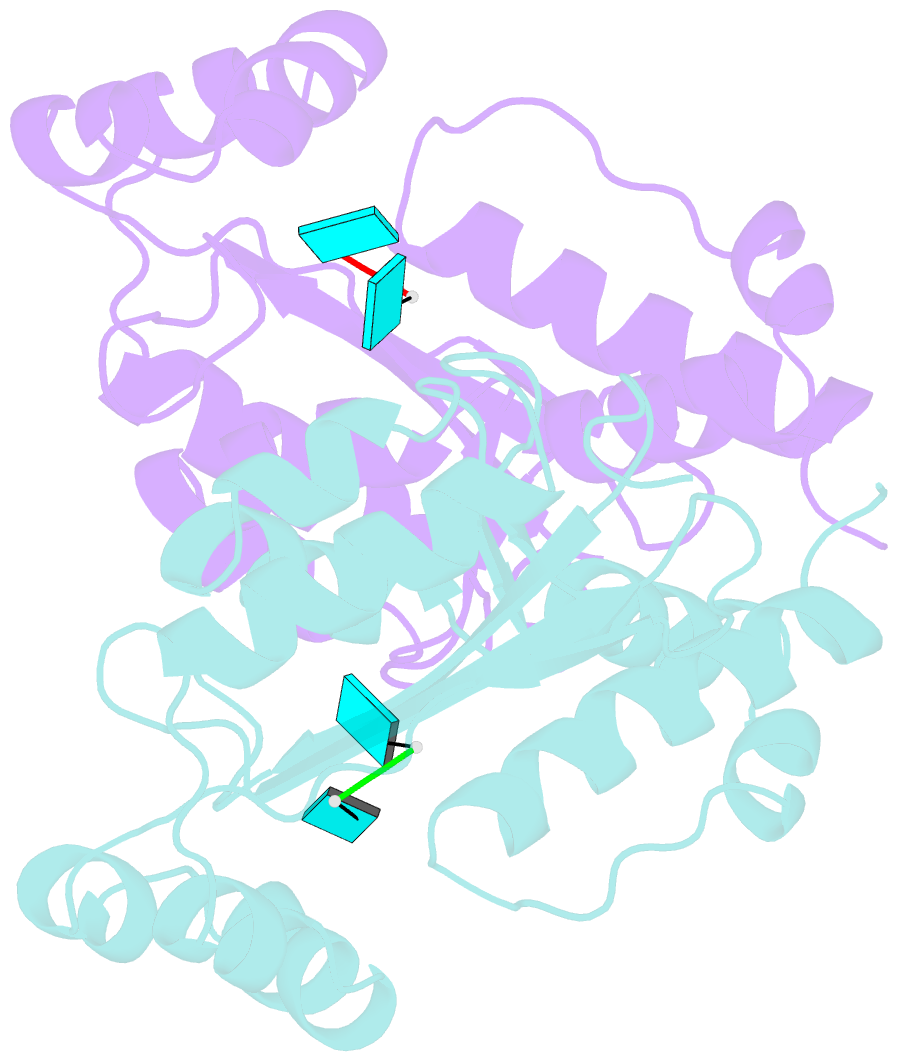
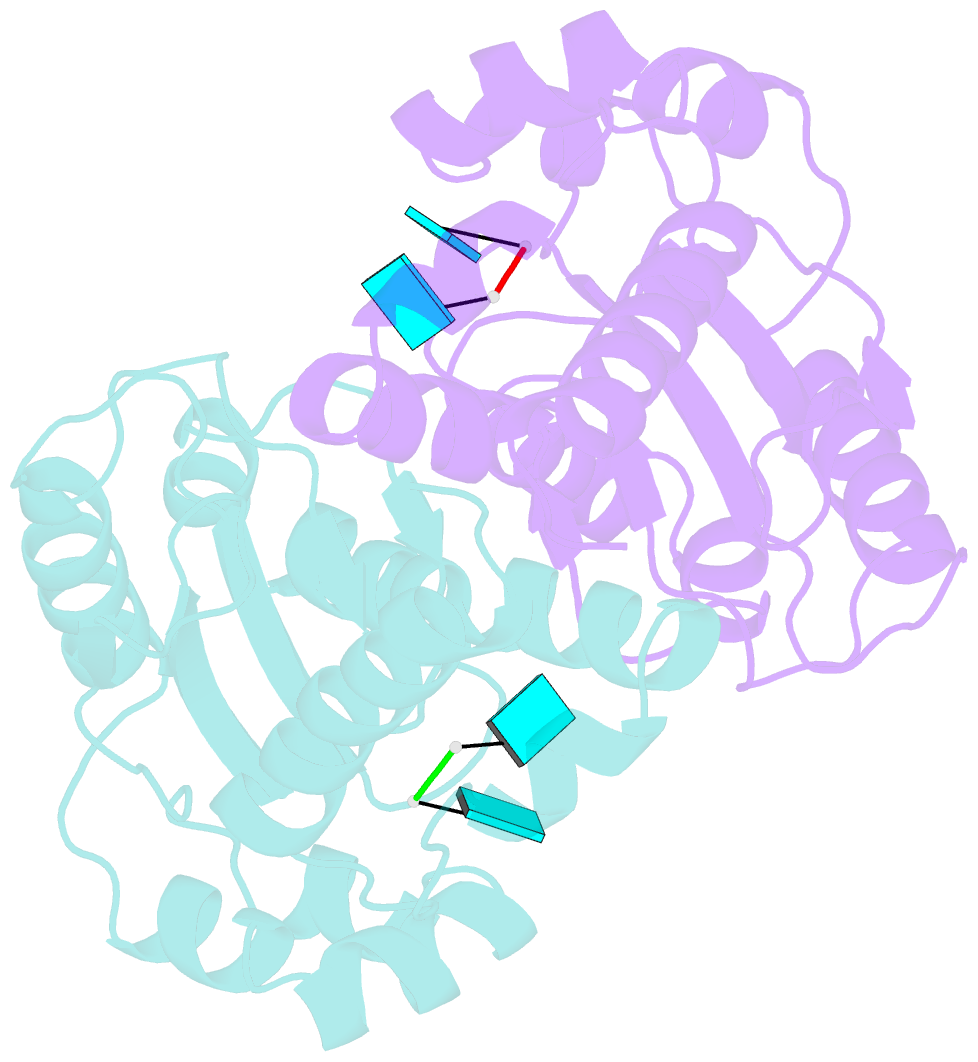
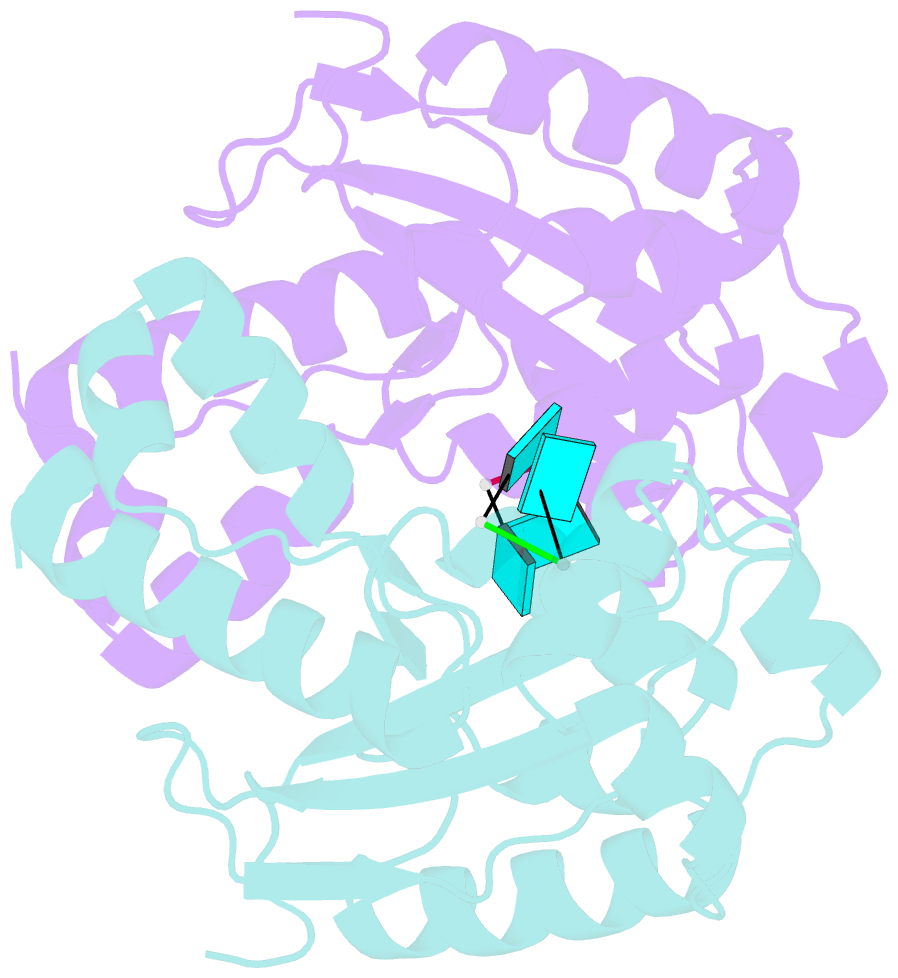
- Two linked uridine bound oligoribonuclease (orn) from colwellia psychrerythraea strain 34h
- Reference
- Lee CW, Park SH, Jeong CS, Cha SS, Park H, Lee JH (2019): "Structural basis of small RNA hydrolysis by oligoribonuclease (CpsORN) from Colwellia psychrerythraea strain 34H." Sci Rep, 9, 2649. doi: 10.1038/s41598-019-39641-0.
- Abstract
- Cells regulate their intracellular mRNA levels by using specific ribonucleases. Oligoribonuclease (ORN) is a 3'-5' exoribonuclease for small RNA molecules, important in RNA degradation and re-utilisation. However, there is no structural information on the ligand-binding form of ORNs. In this study, the crystal structures of oligoribonuclease from Colwellia psychrerythraea strain 34H (CpsORN) were determined in four different forms: unliganded-structure, thymidine 5'-monophosphate p-nitrophenyl ester (pNP-TMP)-bound, two separated uridine-bound, and two linked uridine (U-U)-bound forms. The crystal structures show that CpsORN is a tight dimer, with two separated active sites and one divalent metal cation ion in each active site. These structures represent several snapshots of the enzymatic reaction process, which allowed us to suggest a possible one-metal-dependent reaction mechanism for CpsORN. Moreover, the biochemical data support our suggested mechanism and identified the key residues responsible for enzymatic catalysis of CpsORN.