Summary information and primary citation
- PDB-id
- 6ajo; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- X-ray (2.269 Å)
- Summary
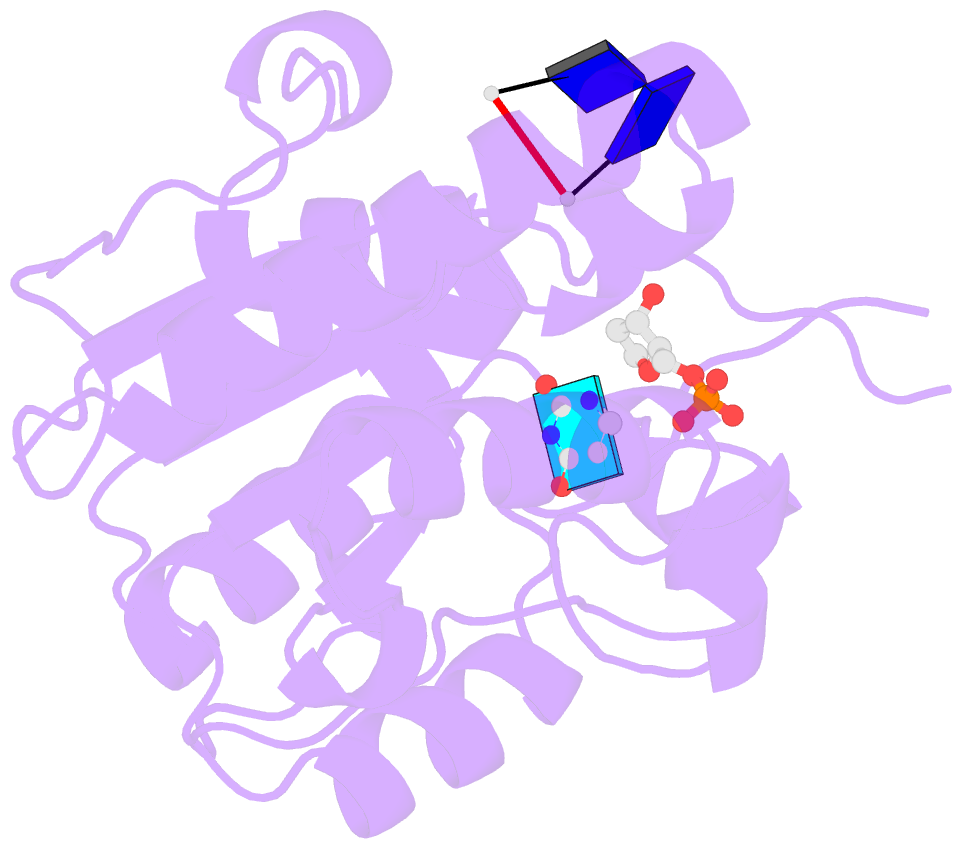
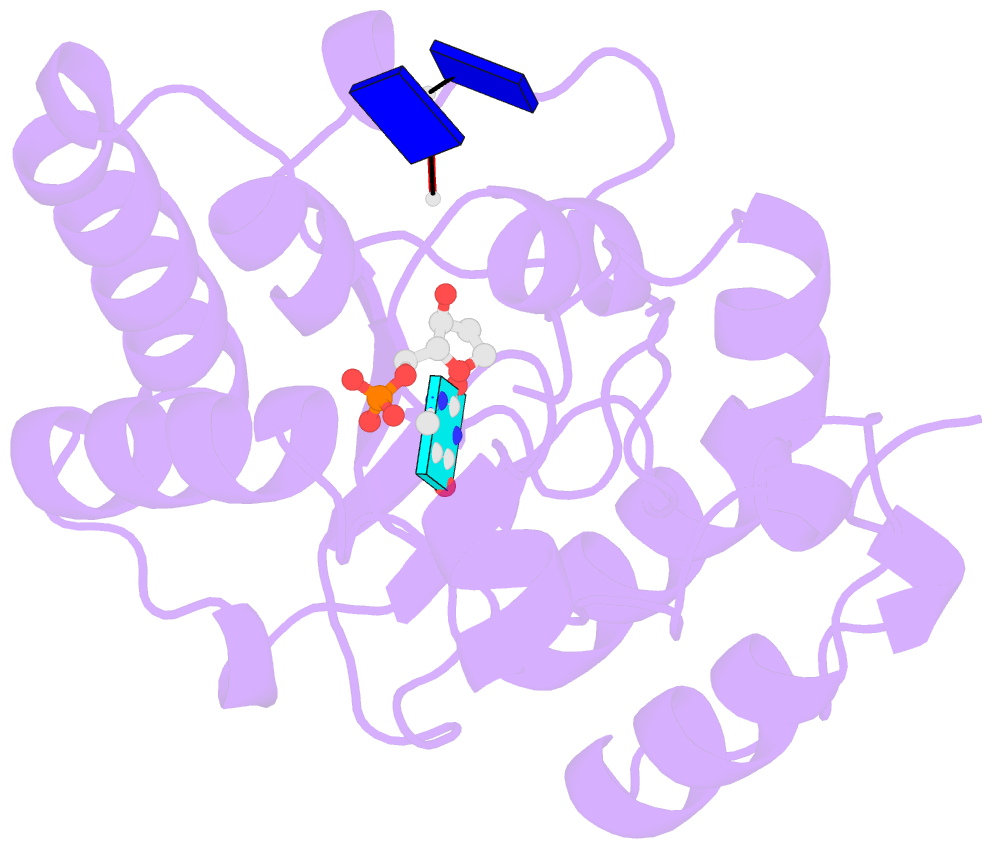
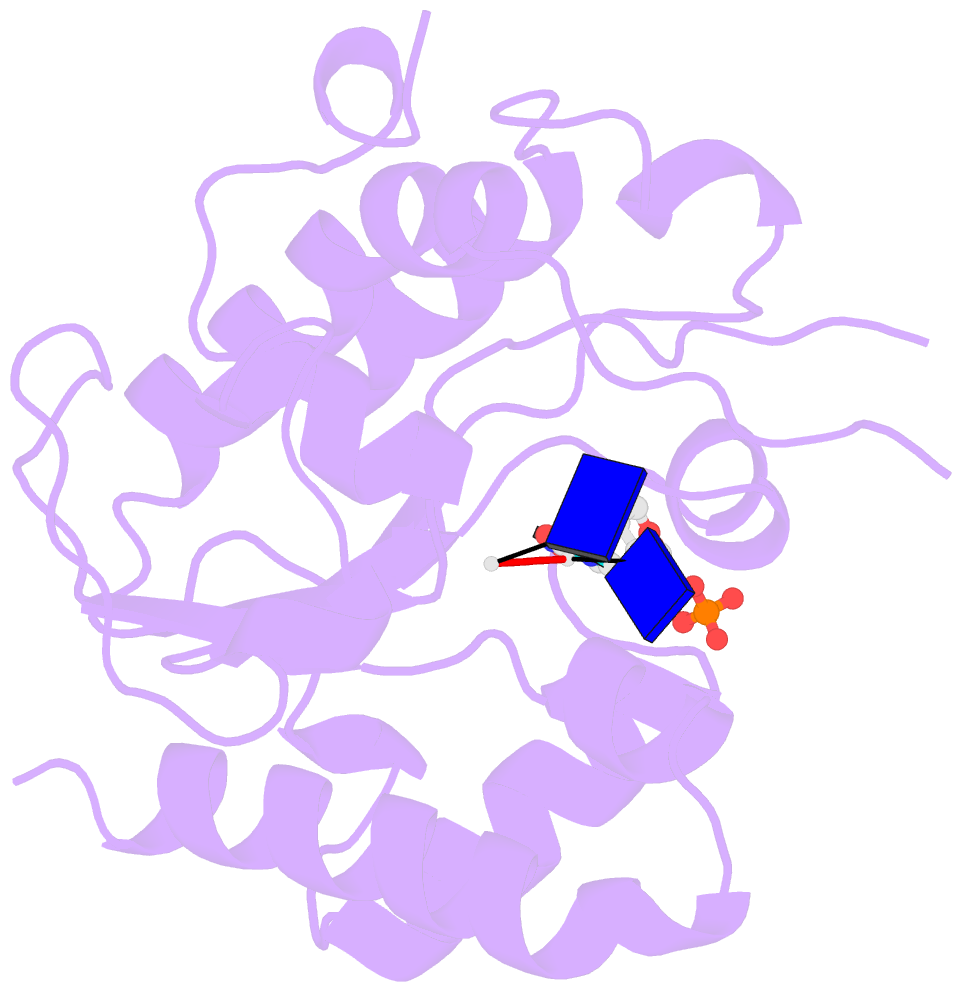
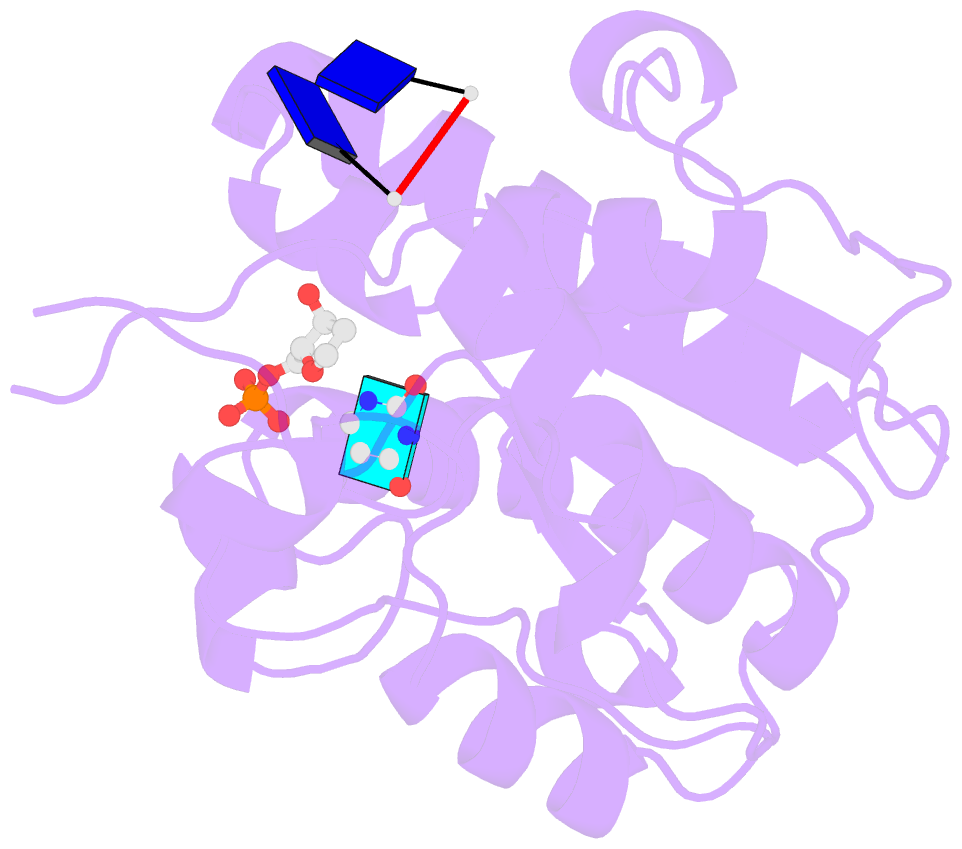
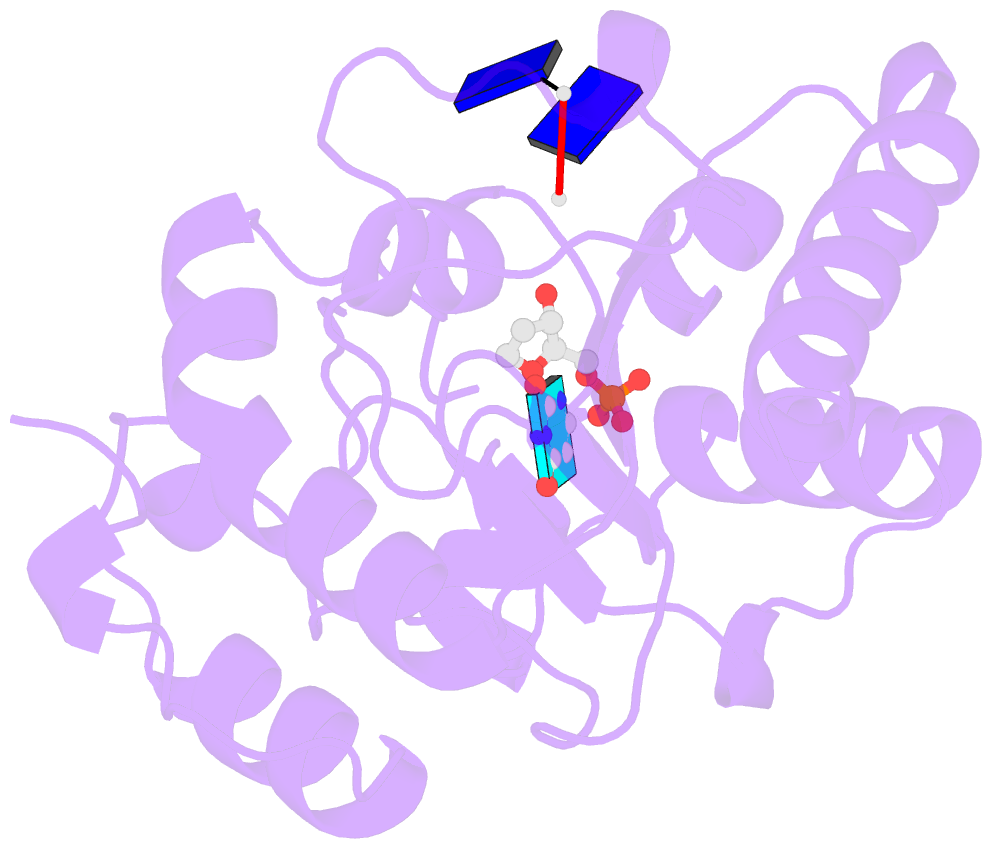
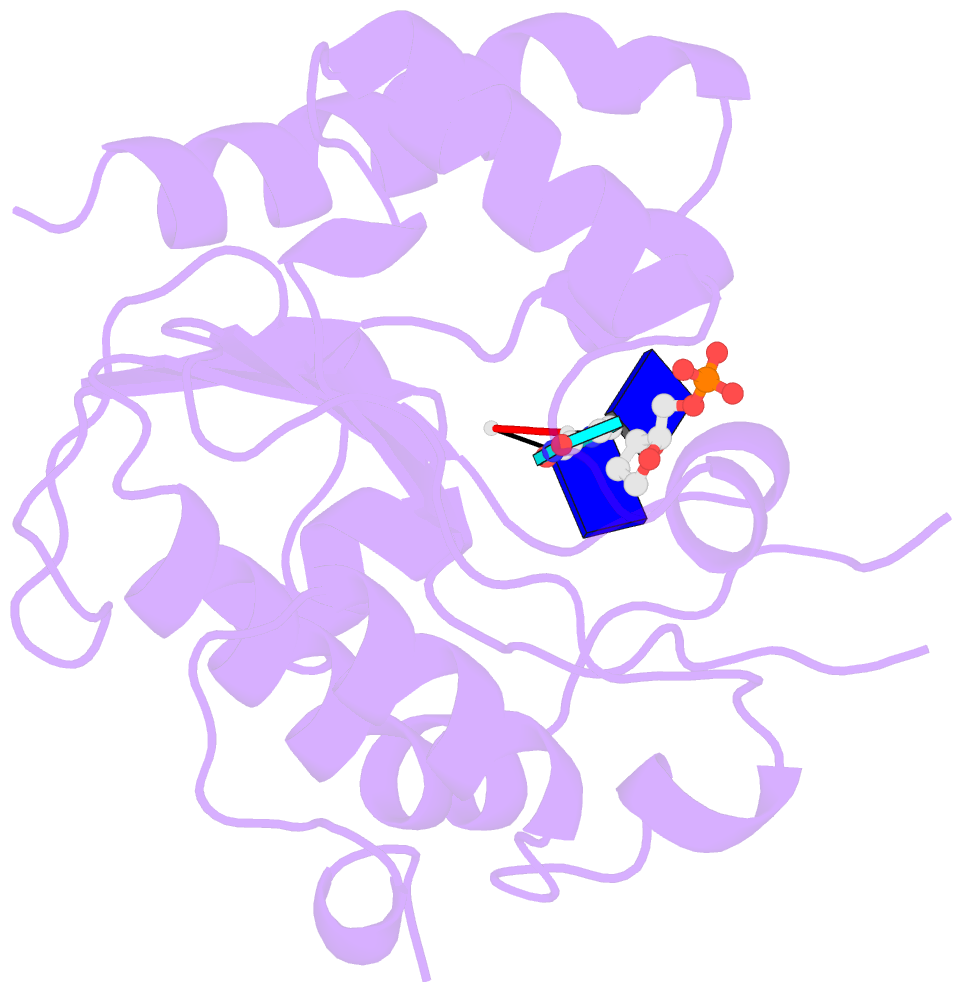
- Complex form of uracil DNA glycosylase x and uracil-DNA.
- Reference
- Ahn WC, Aroli S, Kim JH, Moon JH, Lee GS, Lee MH, Sang PB, Oh BH, Varshney U, Woo EJ (2019): "Covalent binding of uracil DNA glycosylase UdgX to abasic DNA upon uracil excision." Nat.Chem.Biol., 15, 607-614. doi: 10.1038/s41589-019-0289-3.
- Abstract
- Uracil DNA glycosylases (UDGs) are important DNA repair enzymes that excise uracil from DNA, yielding an abasic site. Recently, UdgX, an unconventional UDG with extremely tight binding to DNA containing uracil, was discovered. The structure of UdgX from Mycobacterium smegmatis in complex with DNA shows an overall similarity to that of family 4 UDGs except for a protruding loop at the entrance of the uracil-binding pocket. Surprisingly, H109 in the loop was found to make a covalent bond to the abasic site to form a stable intermediate, while the excised uracil remained in the pocket of the active site. H109 functions as a nucleophile to attack the oxocarbenium ion, substituting for the catalytic water molecule found in other UDGs. To our knowledge, this change from a catalytic water attack to a direct nucleophilic attack by the histidine residue is unprecedented. UdgX utilizes a unique mechanism of protecting cytotoxic abasic sites from exposure to the cellular environment.