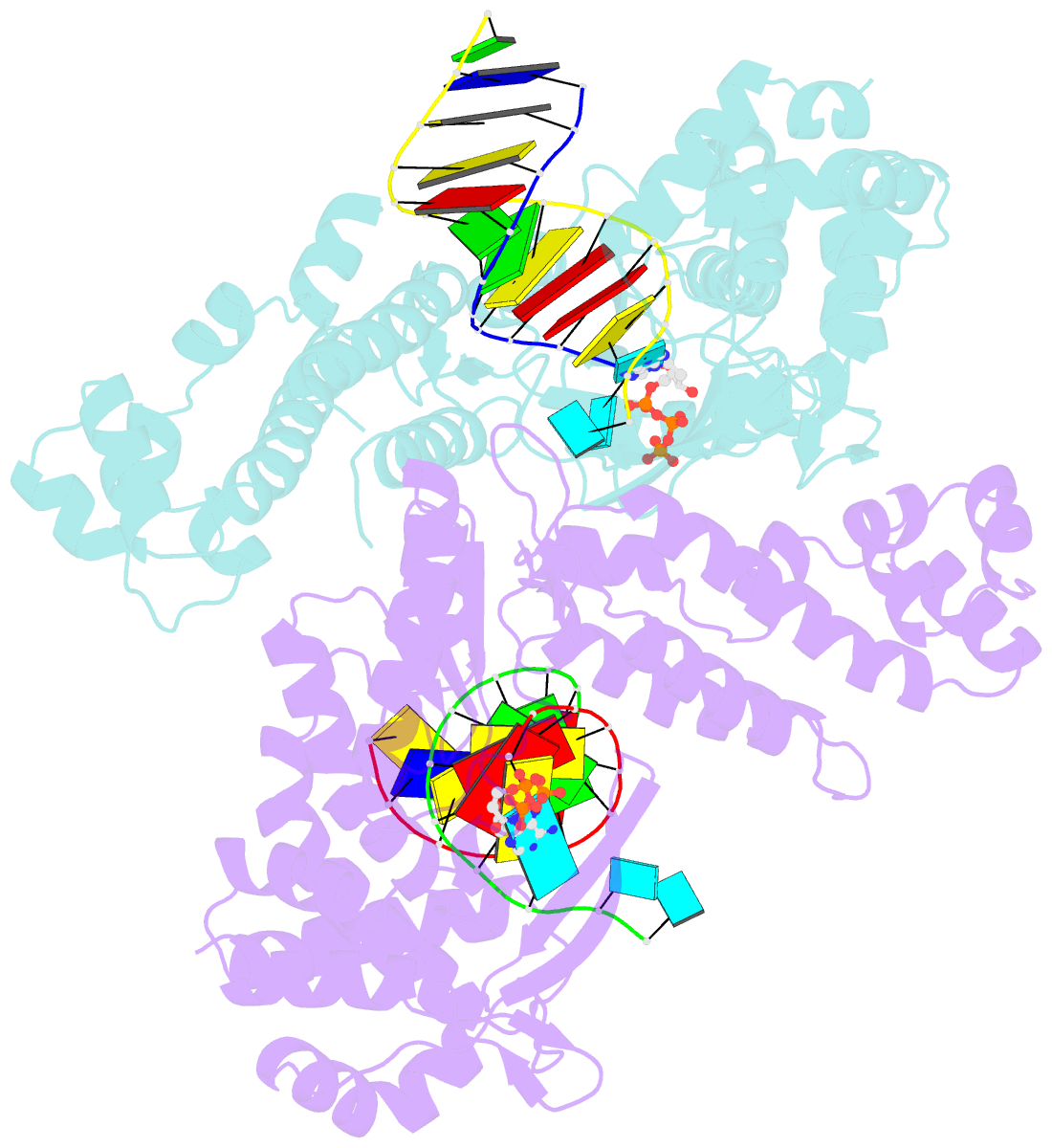
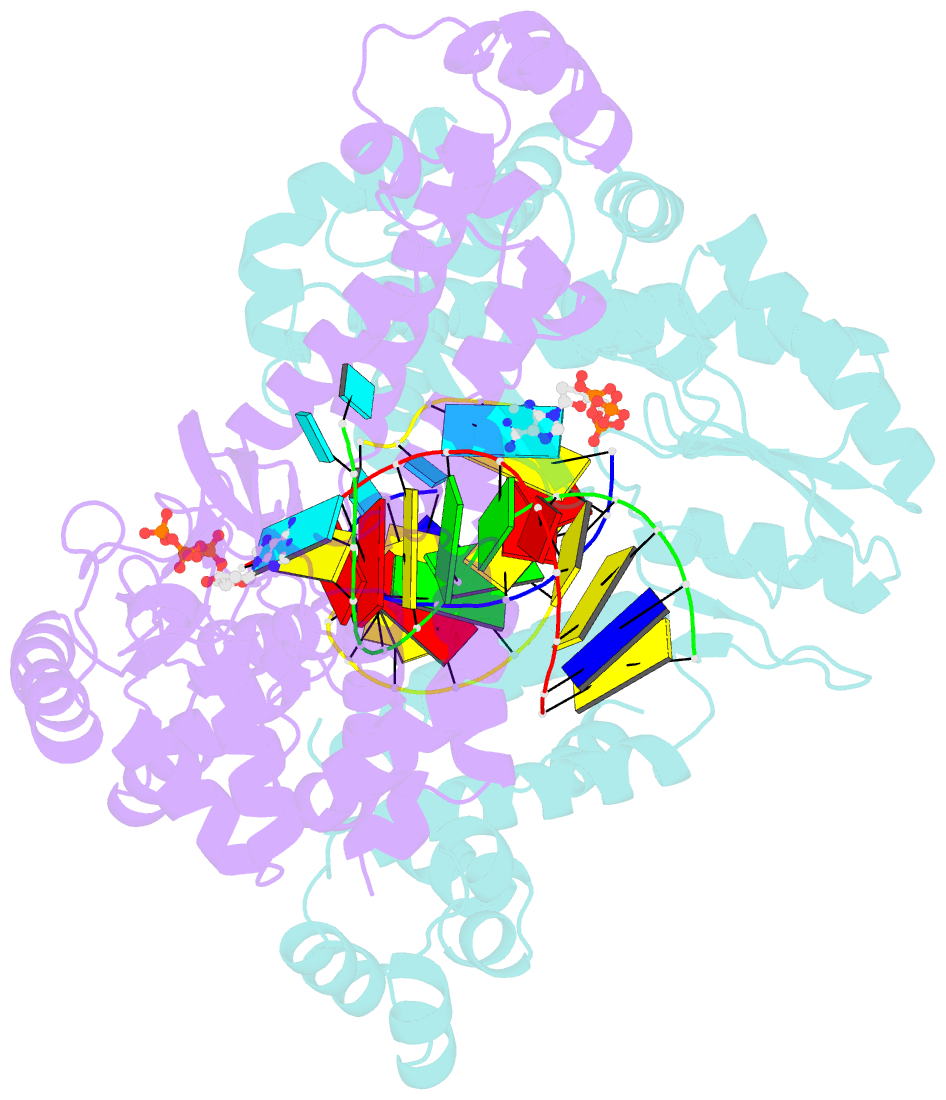
Summary information and primary citation
- PDB-id
- 6ar1; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA-DNA
- Method
- X-ray (3.01 Å)
- Summary
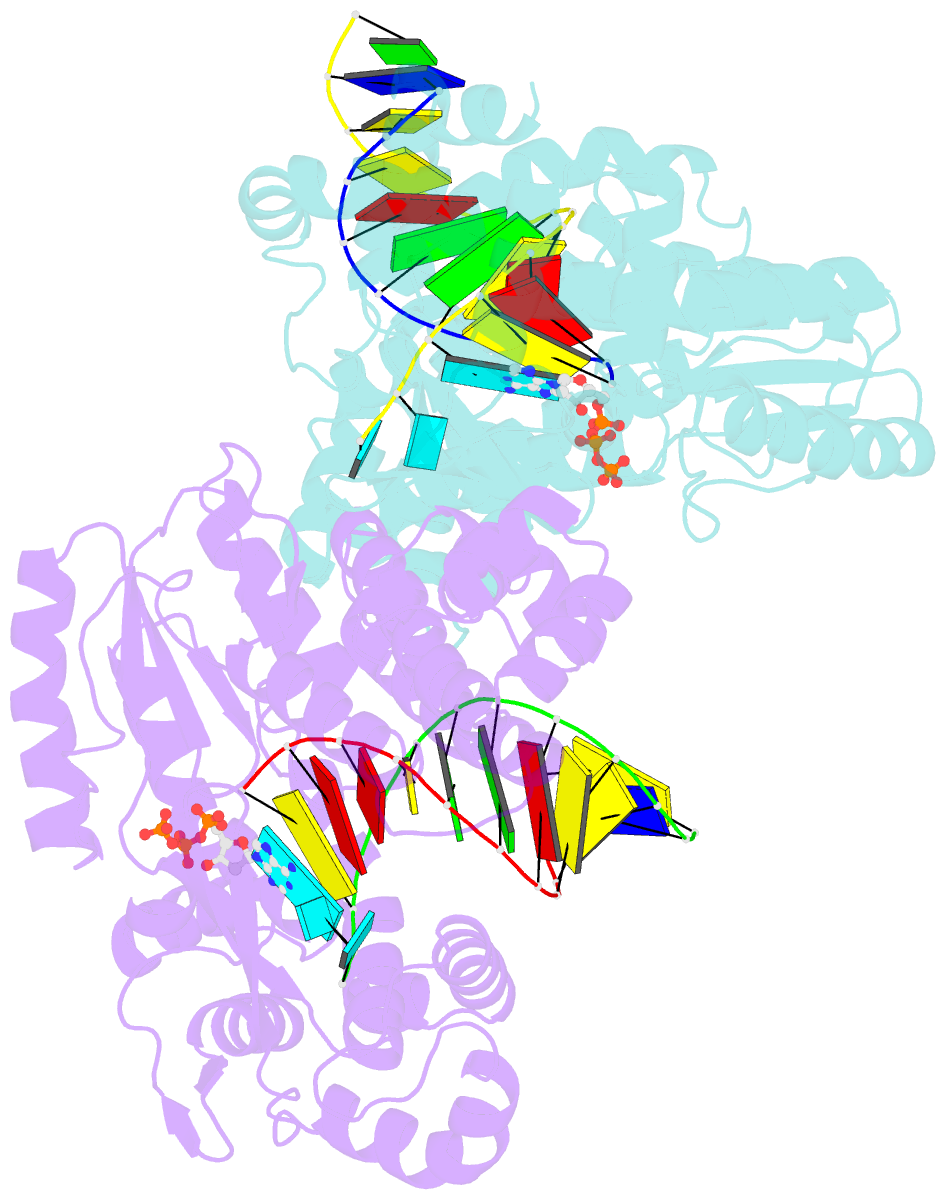
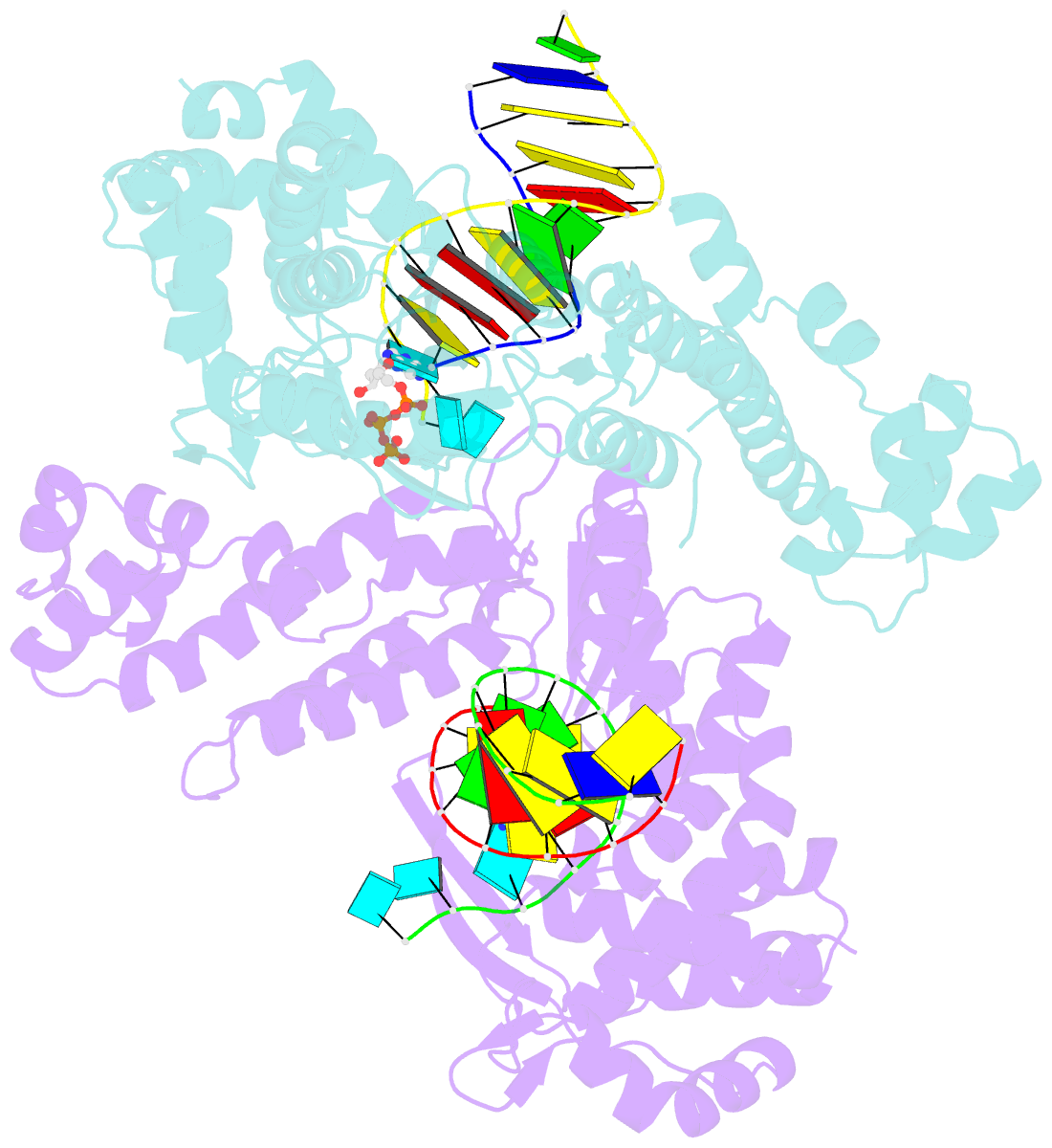
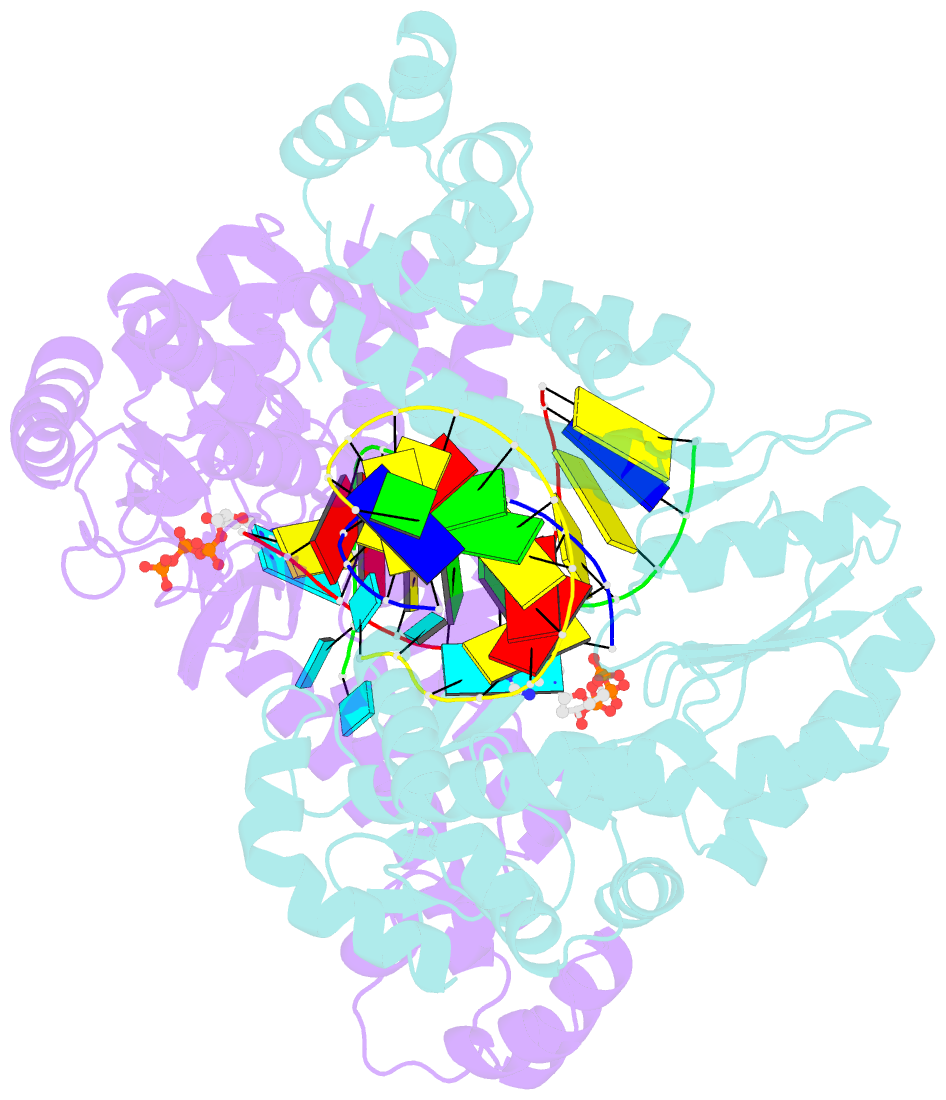
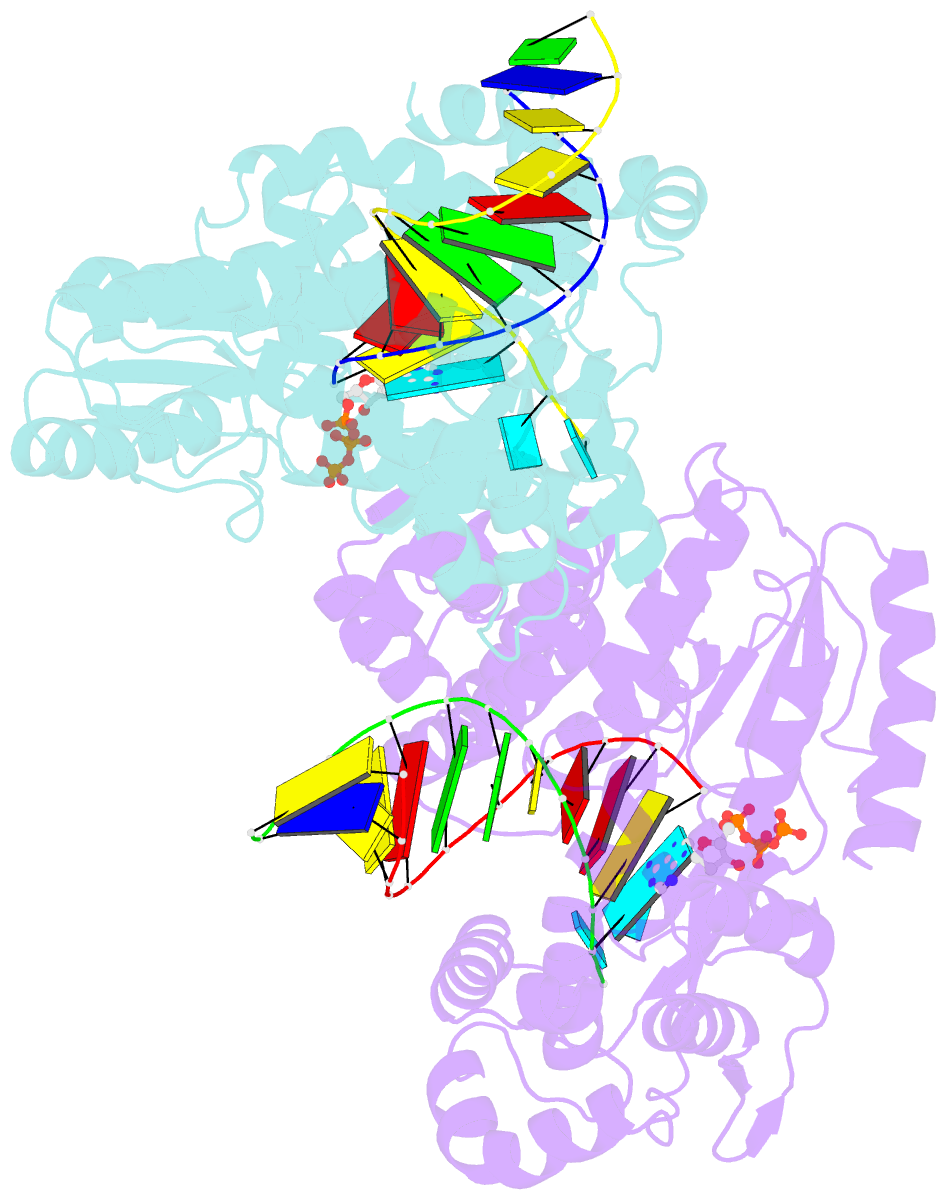
- Structure of a thermostable group ii intron reverse transcriptase with template-primer and its functional and evolutionary implications (rt-duplex (nat))
- Reference
- Stamos JL, Lentzsch AM, Lambowitz AM (2017): "Structure of a Thermostable Group II Intron Reverse Transcriptase with Template-Primer and Its Functional and Evolutionary Implications." Mol. Cell, 68, 926-939.e4. doi: 10.1016/j.molcel.2017.10.024.
- Abstract
- Bacterial group II intron reverse transcriptases (RTs) function in both intron mobility and RNA splicing and are evolutionary predecessors of retrotransposon, telomerase, and retroviral RTs as well as the spliceosomal protein Prp8 in eukaryotes. Here we determined a crystal structure of a full-length thermostable group II intron RT in complex with an RNA template-DNA primer duplex and incoming deoxynucleotide triphosphate (dNTP) at 3.0-Å resolution. We find that the binding of template-primer and key aspects of the RT active site are surprisingly different from retroviral RTs but remarkably similar to viral RNA-dependent RNA polymerases. The structure reveals a host of features not seen previously in RTs that may contribute to distinctive biochemical properties of group II intron RTs, and it provides a prototype for many related bacterial and eukaryotic non-LTR retroelement RTs. It also reveals how protein structural features used for reverse transcription evolved to promote the splicing of both group II and spliceosomal introns.