Summary information and primary citation
- PDB-id
- 6as7; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- replication-DNA
- Method
- X-ray (2.95 Å)
- Summary
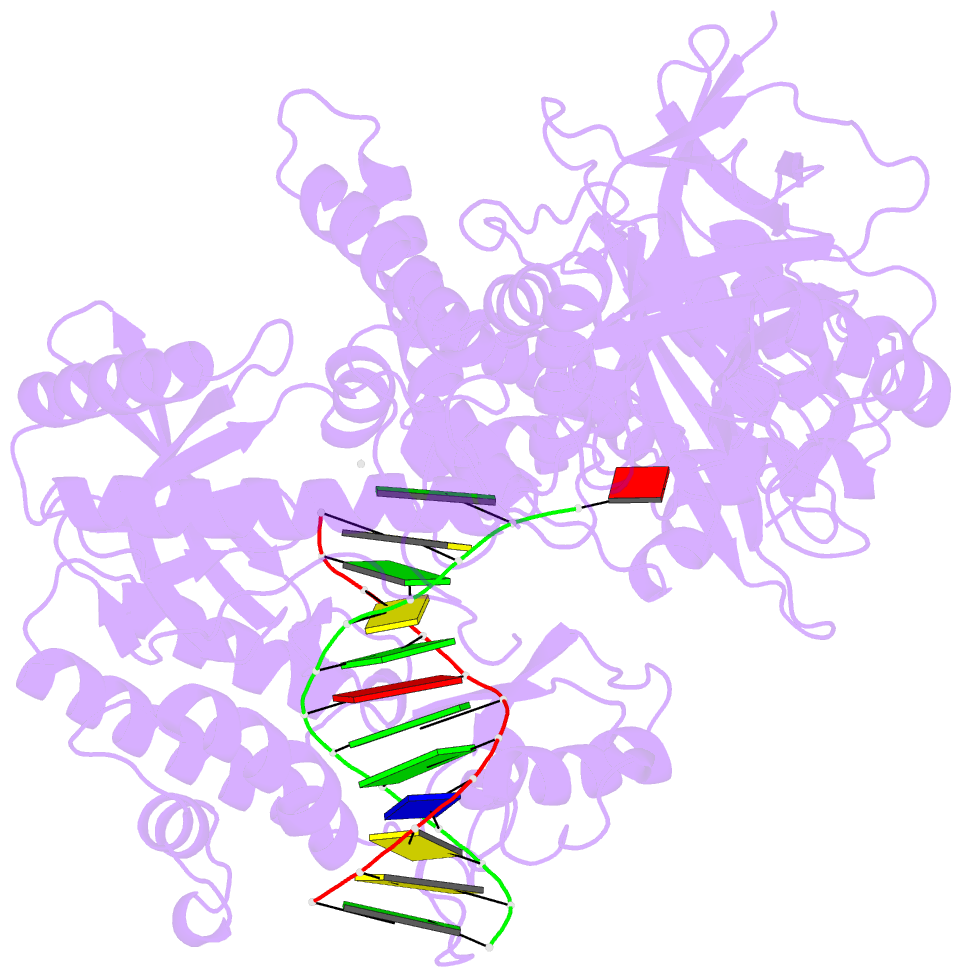
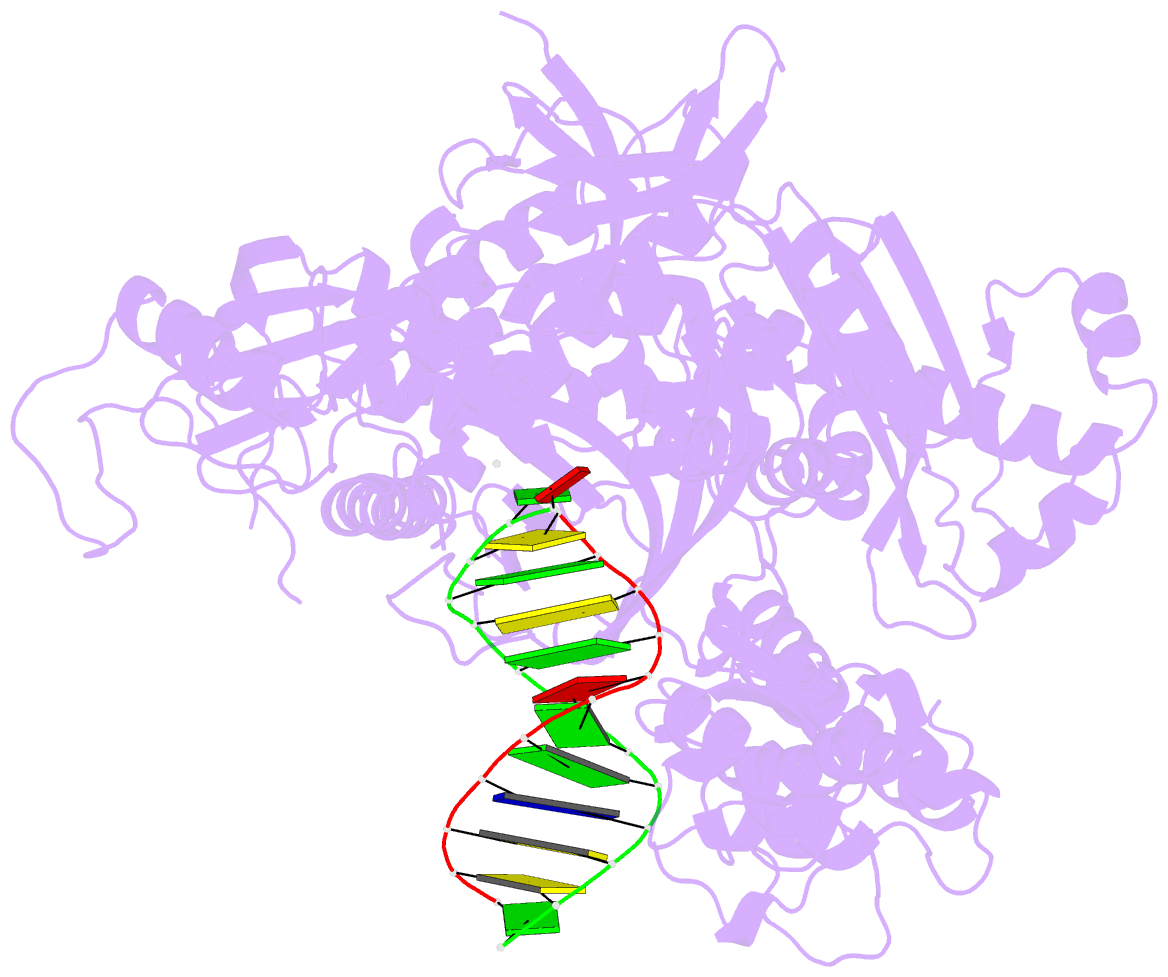
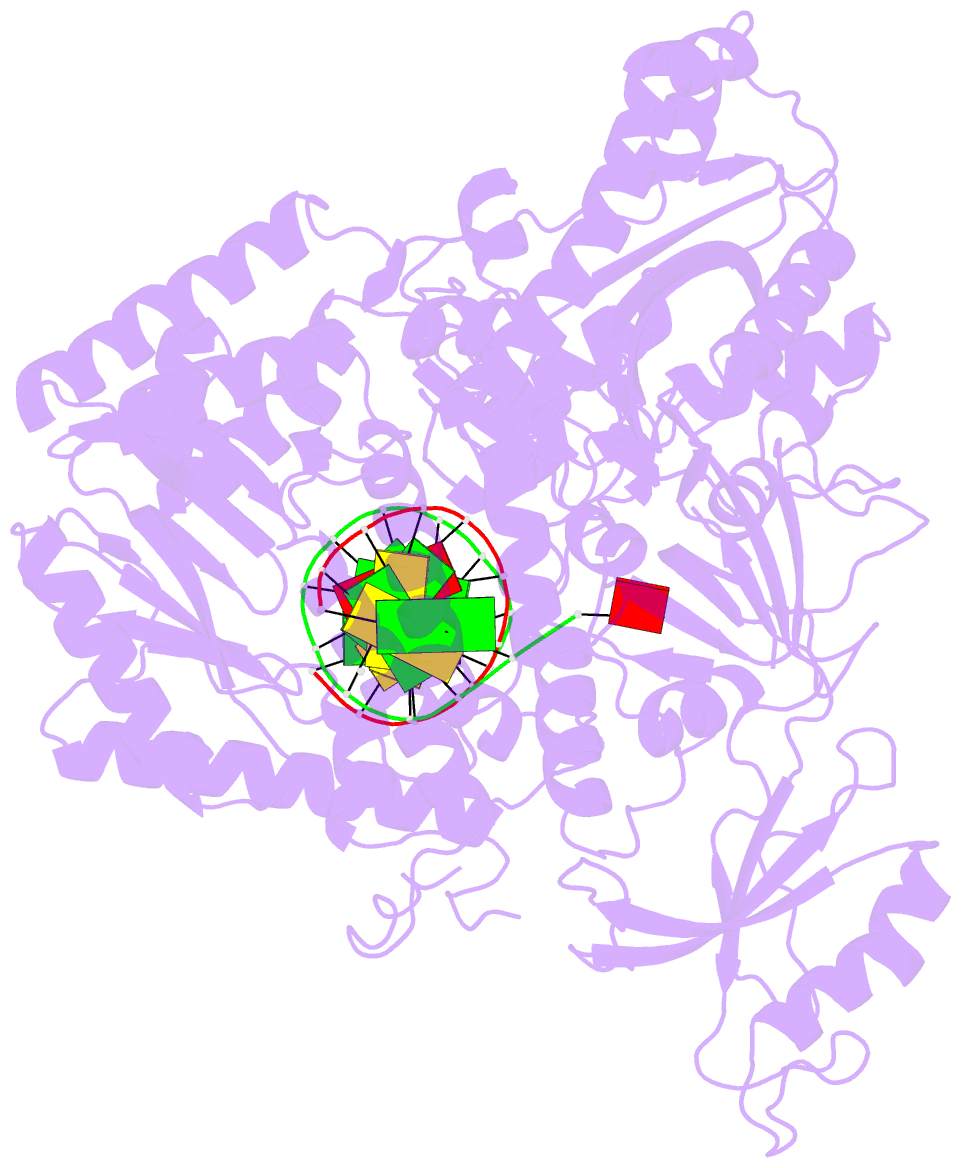
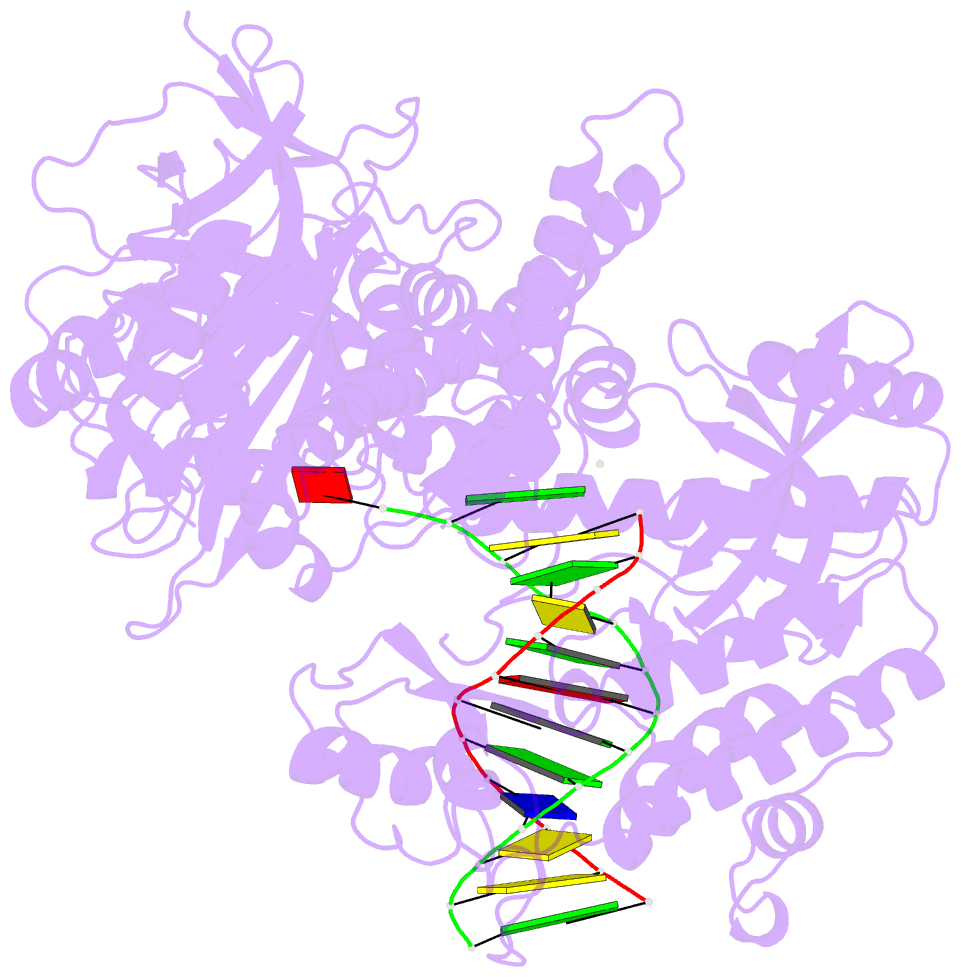
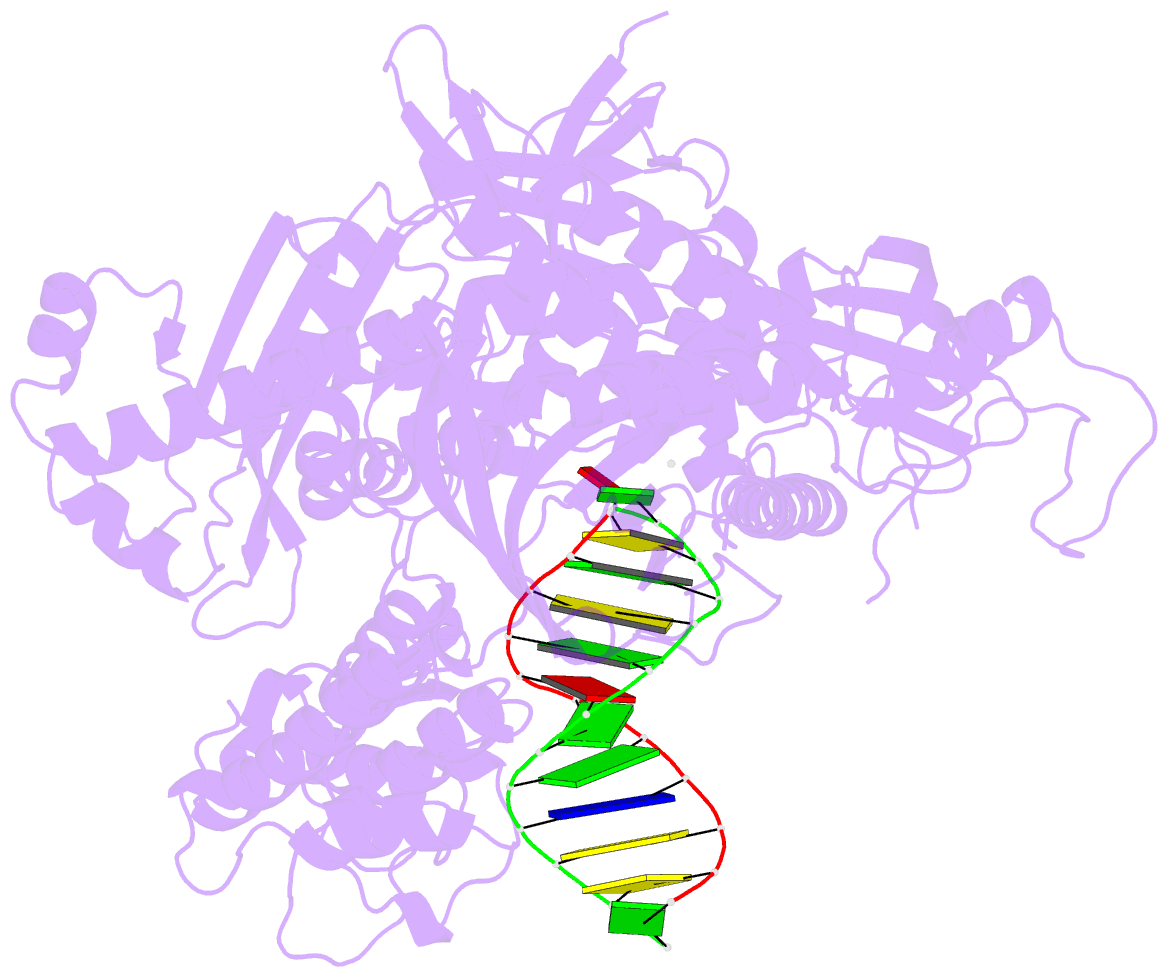
- Crystal structure of the catalytic core of human DNA polymerase alpha in ternary complex with an DNA-primed DNA template and dctp
- Reference
- Baranovskiy AG, Duong VN, Babayeva ND, Zhang Y, Pavlov YI, Anderson KS, Tahirov TH (2018): "Activity and fidelity of human DNA polymerase alpha depend on primer structure." J. Biol. Chem., 293, 6824-6843. doi: 10.1074/jbc.RA117.001074.
- Abstract
- DNA polymerase α (Polα) plays an important role in genome replication. In a complex with primase, Polα synthesizes chimeric RNA-DNA primers necessary for replication of both chromosomal DNA strands. During RNA primer extension with deoxyribonucleotides, Polα needs to use double-stranded helical substrates having different structures. Here, we provide a detailed structure-function analysis of human Polα's interaction with dNTPs and DNA templates primed with RNA, chimeric RNA-DNA, or DNA. We report the crystal structures of two ternary complexes of the Polα catalytic domain containing dCTP, a DNA template, and either a DNA or an RNA primer. Unexpectedly, in the ternary complex with a DNA:DNA duplex and dCTP, the "fingers" subdomain of Polα is in the open conformation. Polα induces conformational changes in the DNA and hybrid duplexes to produce the universal double helix form. Pre-steady-state kinetic studies indicated for both duplex types that chemical catalysis rather than product release is the rate-limiting step. Moreover, human Polα extended DNA primers with higher efficiency but lower processivity than it did with RNA and chimeric primers. Polα has a substantial propensity to make errors during DNA synthesis, and we observed that its fidelity depends on the type of sugar at the primer 3'-end. A detailed structural comparison of Polα with other replicative DNA polymerases disclosed common features and some differences, which may reflect the specialization of each polymerase in genome replication.