Summary information and primary citation
- PDB-id
- 6bbo; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-RNA
- Method
- X-ray (3.428 Å)
- Summary
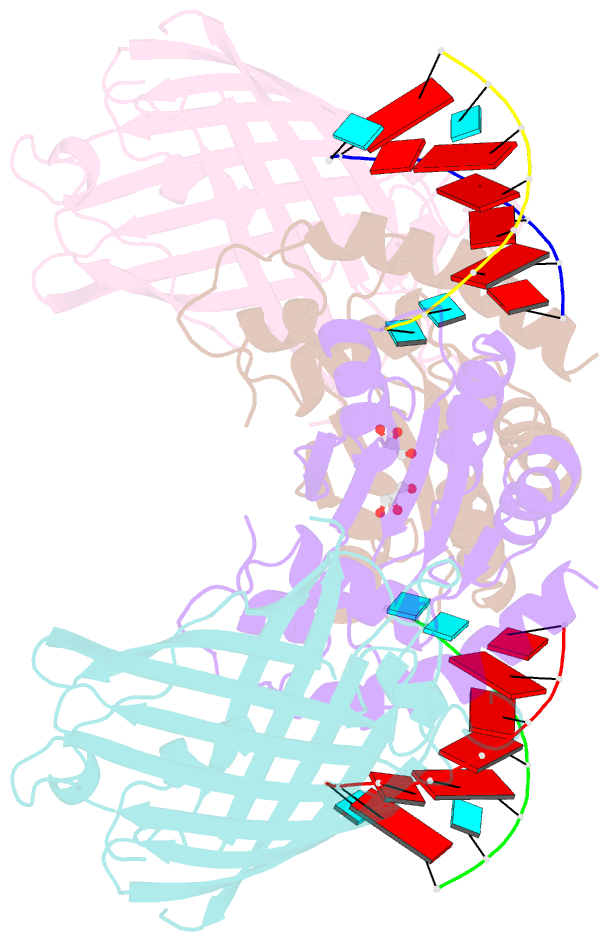
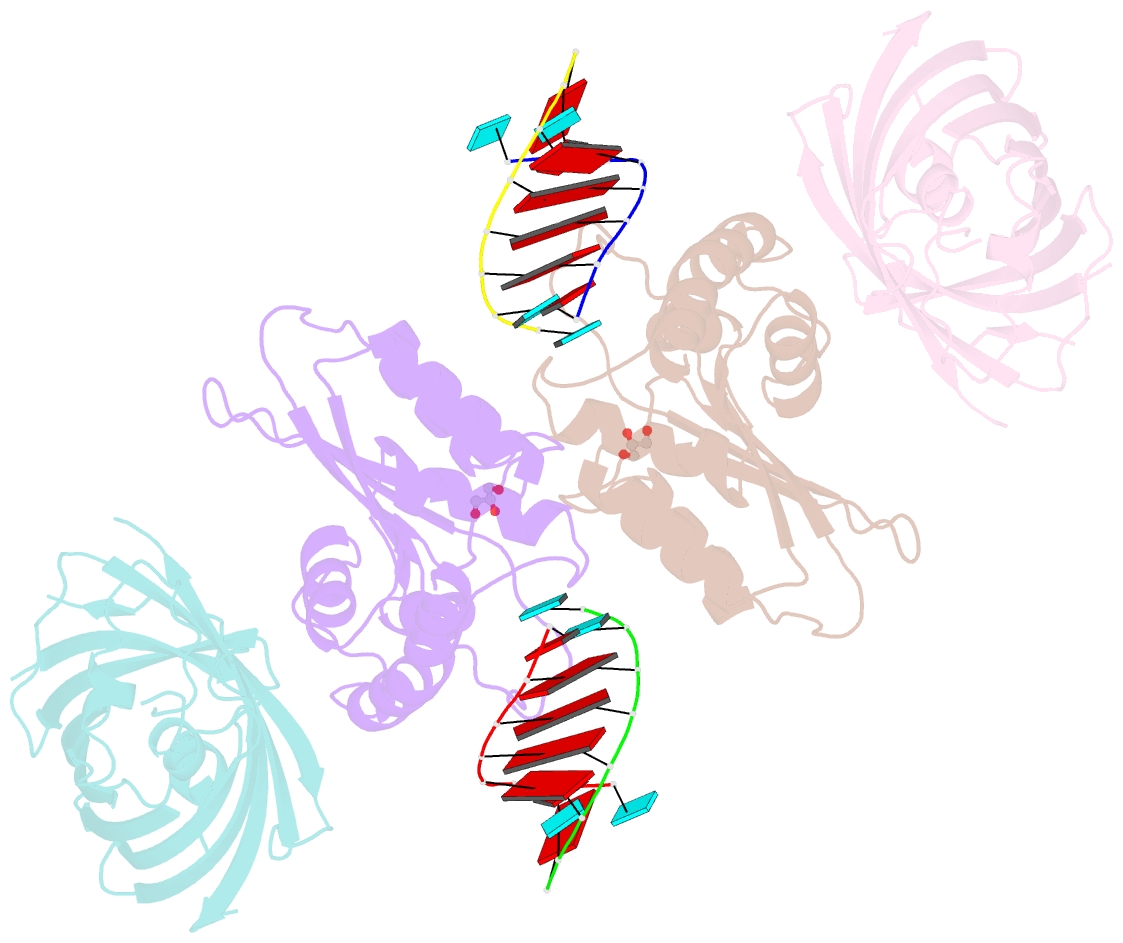
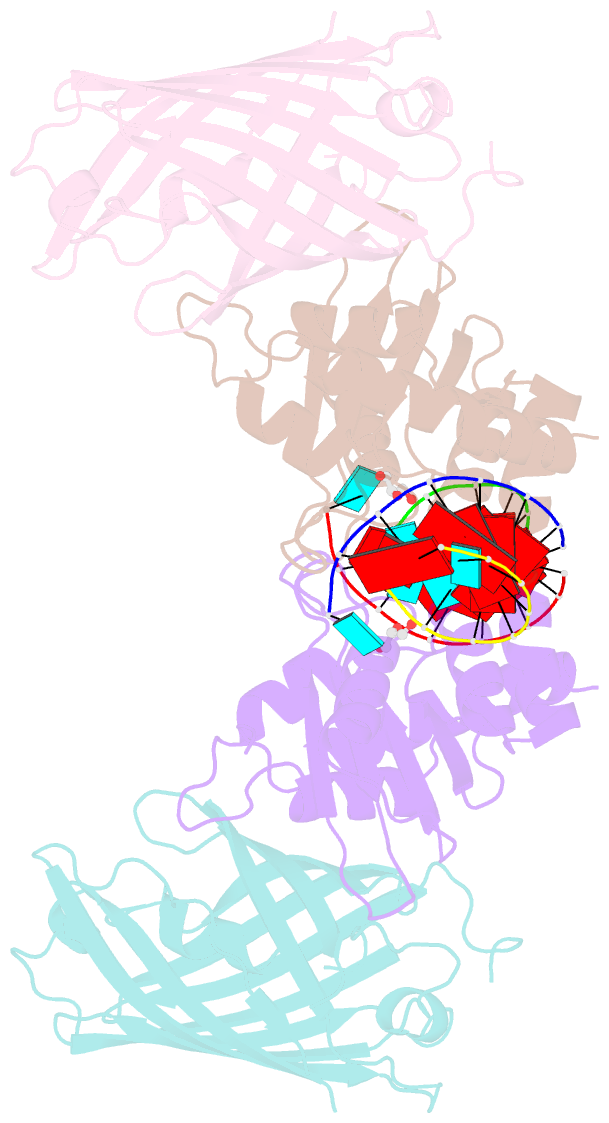
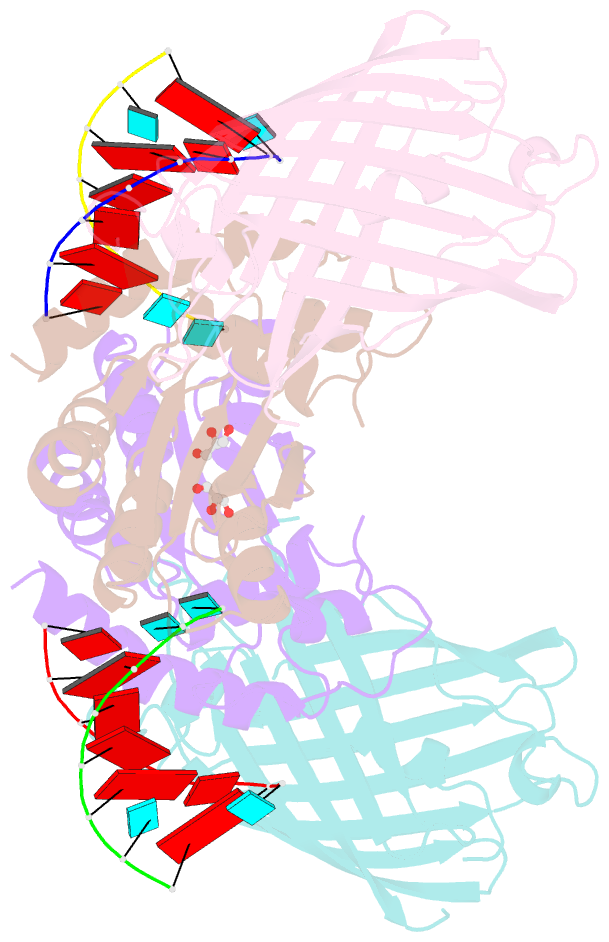
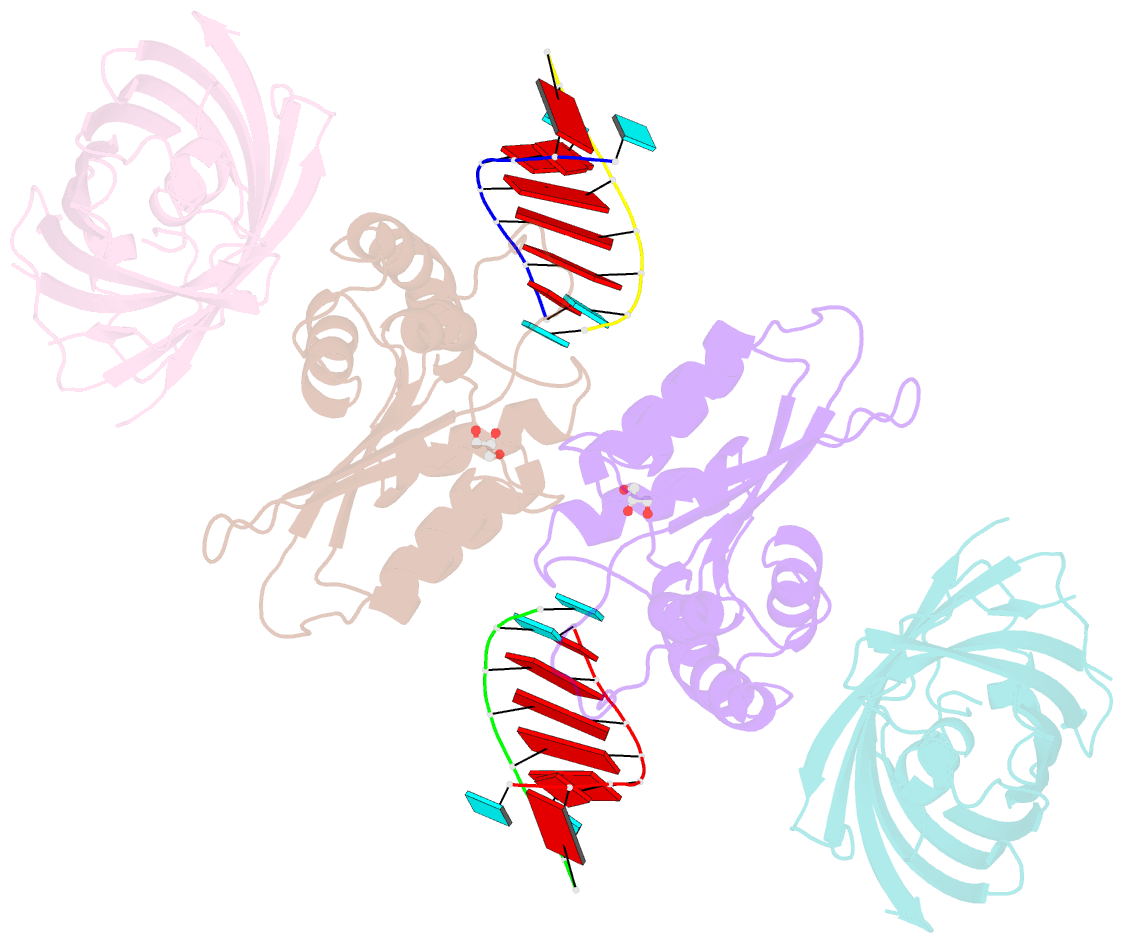
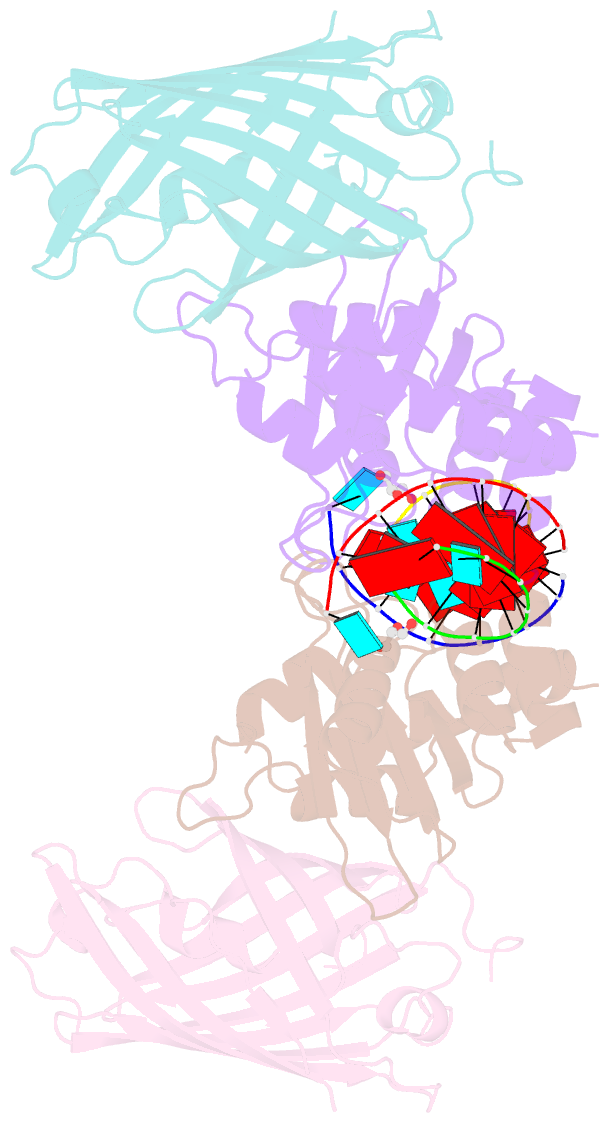
- Crystal structure of human apobec3h-RNA complex
- Reference
- Shaban NM, Shi K, Lauer KV, Carpenter MA, Richards CM, Salamango D, Wang J, Lopresti MW, Banerjee S, Levin-Klein R, Brown WL, Aihara H, Harris RS (2018): "The Antiviral and Cancer Genomic DNA Deaminase APOBEC3H Is Regulated by an RNA-Mediated Dimerization Mechanism." Mol. Cell, 69, 75-86.e9. doi: 10.1016/j.molcel.2017.12.010.
- Abstract
- Human APOBEC3H and homologous single-stranded DNA cytosine deaminases are unique to mammals. These DNA-editing enzymes function in innate immunity by restricting the replication of viruses and transposons. APOBEC3H also contributes to cancer mutagenesis. Here, we address the fundamental nature of RNA in regulating human APOBEC3H activities. APOBEC3H co-purifies with RNA as an inactive protein, and RNase A treatment enables strong DNA deaminase activity. RNA-binding-defective mutants demonstrate clear separation of function by becoming DNA hypermutators. Biochemical and crystallographic data demonstrate a mechanism in which double-stranded RNA mediates enzyme dimerization. Additionally, APOBEC3H separation-of-function mutants show that RNA binding is required for cytoplasmic localization, packaging into HIV-1 particles, and antiviral activity. Overall, these results support a model in which structured RNA negatively regulates the potentially harmful DNA deamination activity of APOBEC3H while, at the same time, positively regulating its antiviral activity.