Summary information and primary citation
- PDB-id
- 6bux; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (1.856 Å)
- Summary
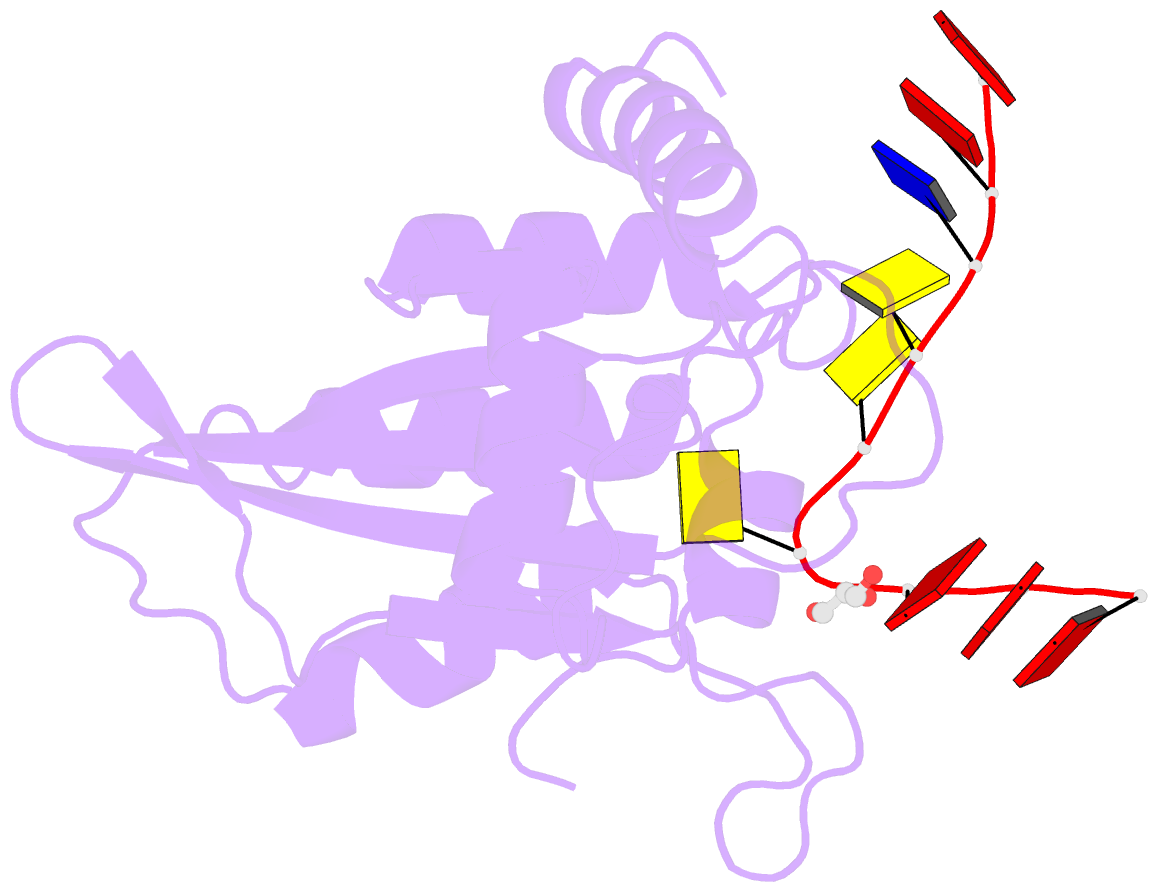
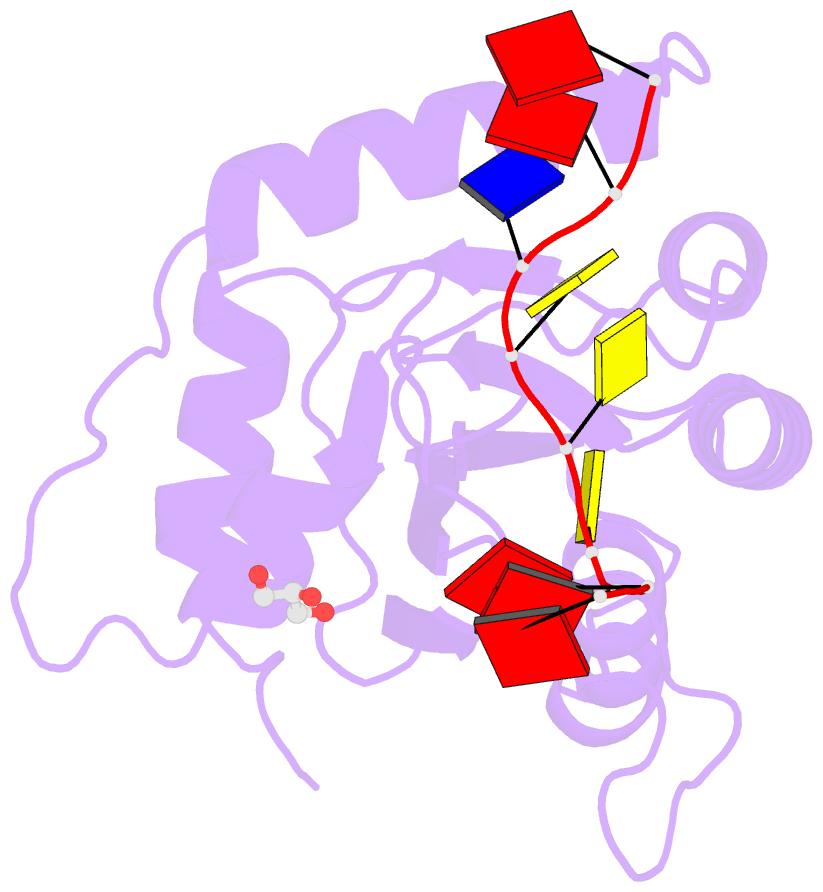
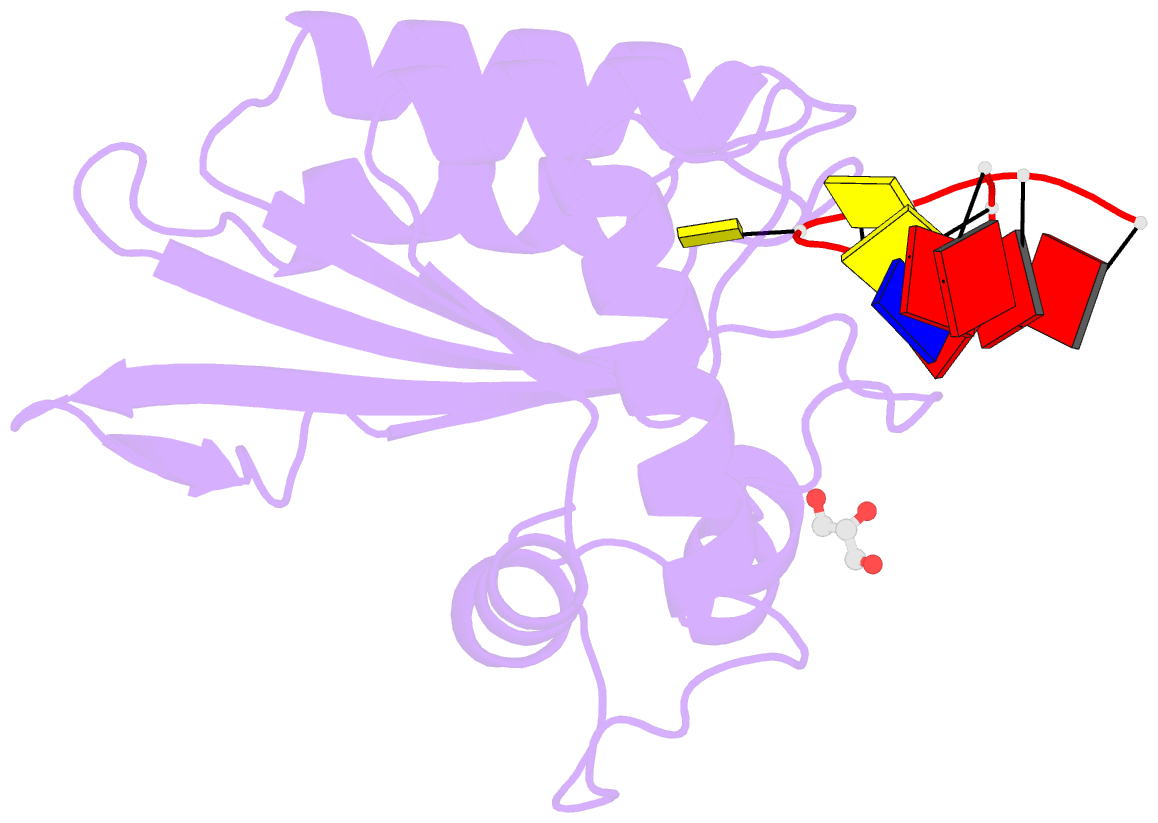
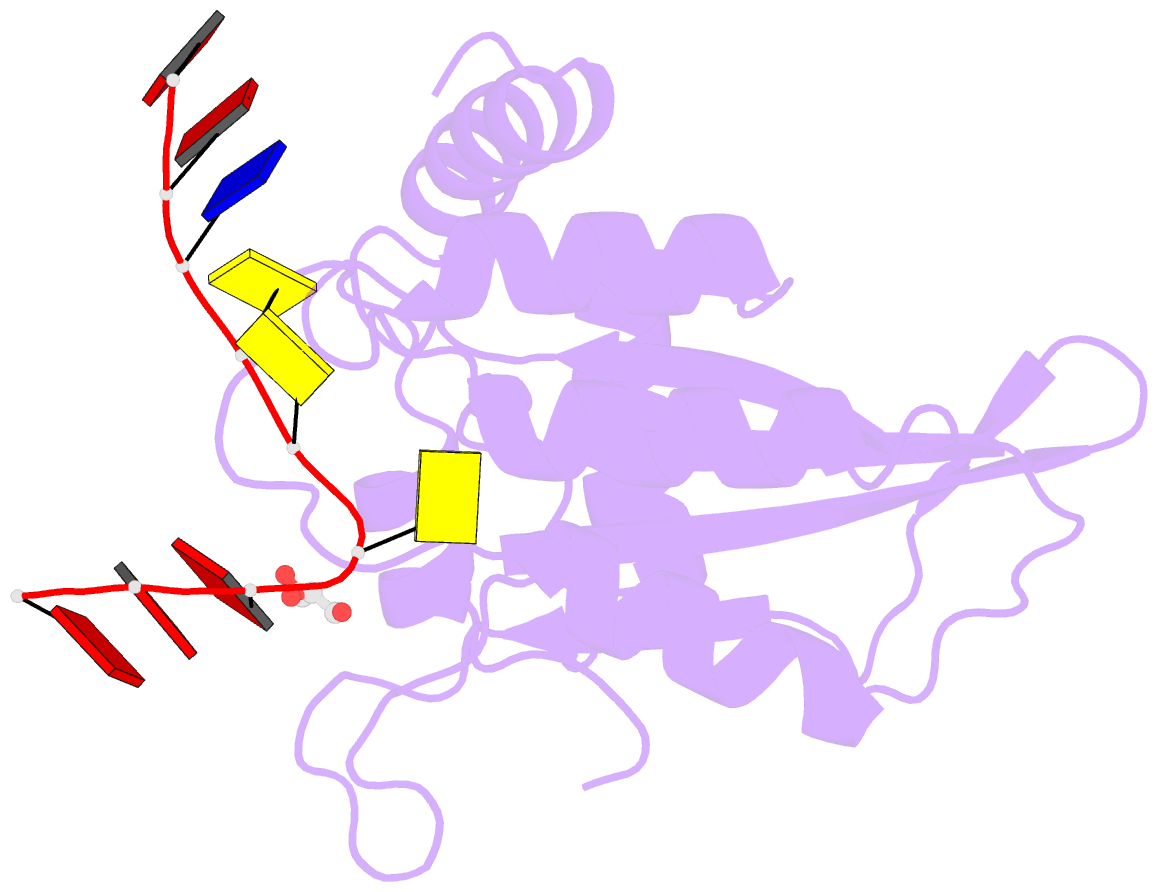
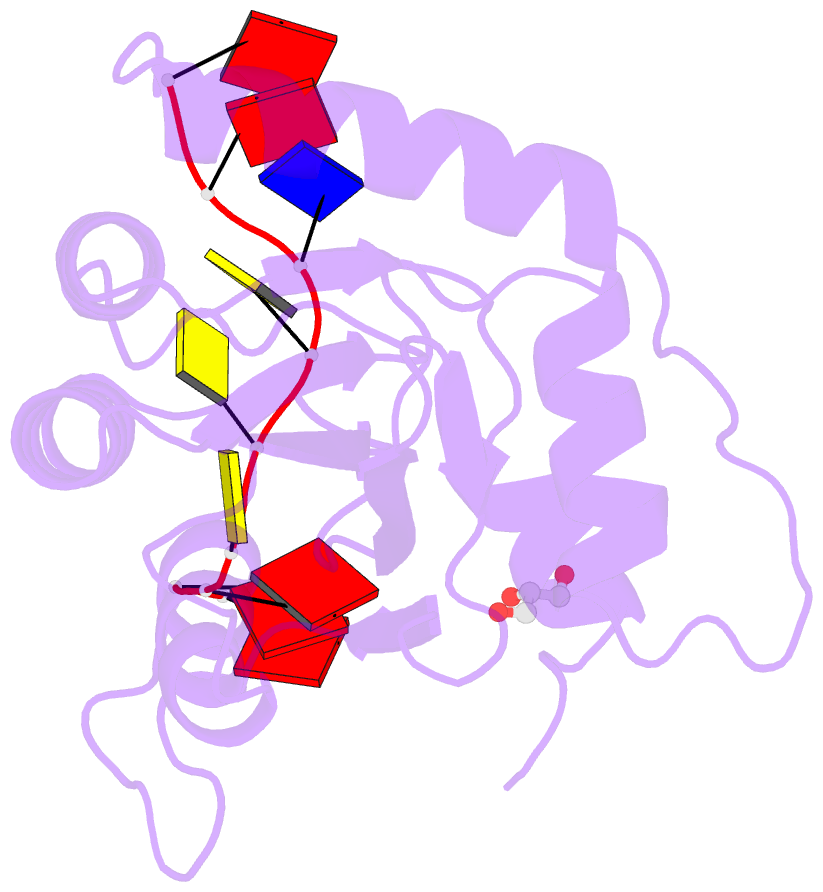
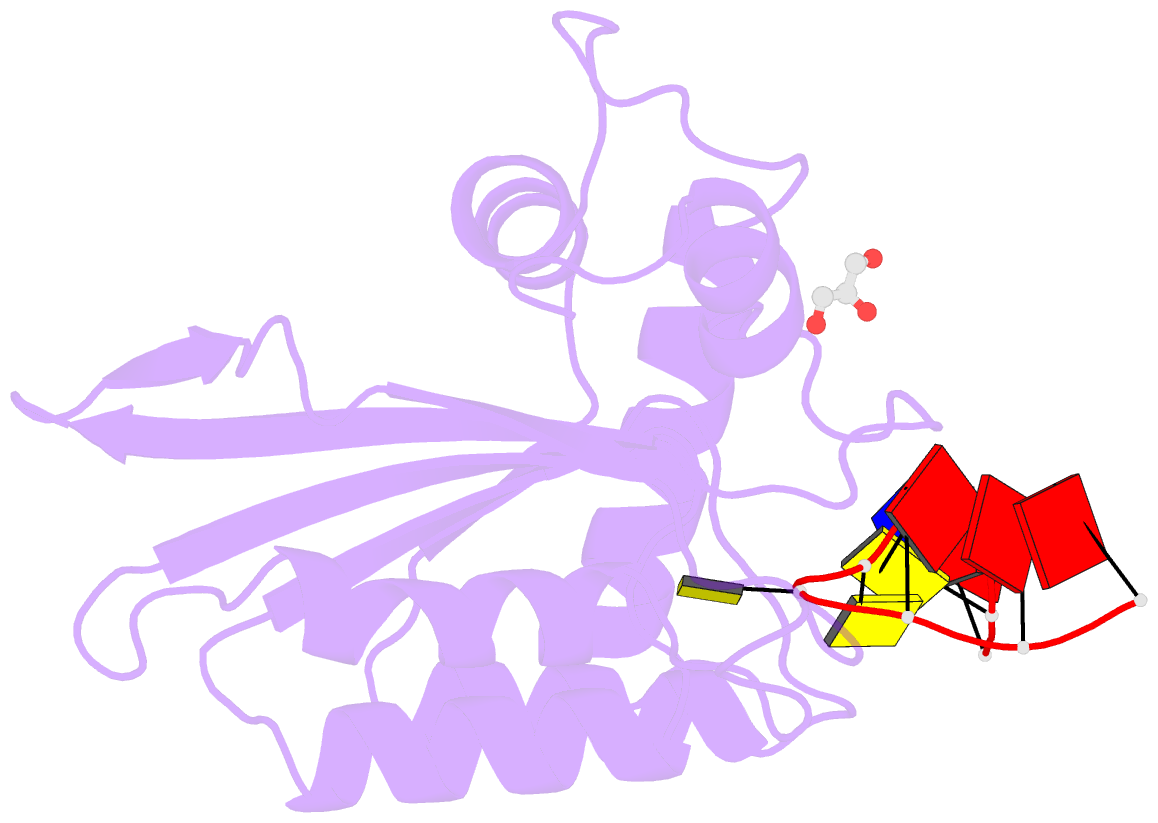
- Crystal structure of apobec3g catalytic domain complex with substrate ssDNA
- Reference
- Maiti A, Myint W, Kanai T, Delviks-Frankenberry K, Sierra Rodriguez C, Pathak VK, Schiffer CA, Matsuo H (2018): "Crystal structure of the catalytic domain of HIV-1 restriction factor APOBEC3G in complex with ssDNA." Nat Commun, 9, 2460. doi: 10.1038/s41467-018-04872-8.
- Abstract
- The human APOBEC3G protein is a cytidine deaminase that generates cytidine to deoxy-uridine mutations in single-stranded DNA (ssDNA), and capable of restricting replication of HIV-1 by generating mutations in viral genome. The mechanism by which APOBEC3G specifically deaminates 5'-CC motifs has remained elusive since structural studies have been hampered due to apparently weak ssDNA binding of the catalytic domain of APOBEC3G. We overcame the problem by generating a highly active variant with higher ssDNA affinity. Here, we present the crystal structure of this variant complexed with a ssDNA substrate at 1.86 Å resolution. This structure reveals atomic-level interactions by which APOBEC3G recognizes a functionally-relevant 5'-TCCCA sequence. This complex also reveals a key role of W211 in substrate recognition, implicating a similar recognition in activation-induced cytidine deaminase (AID) with a conserved tryptophan.