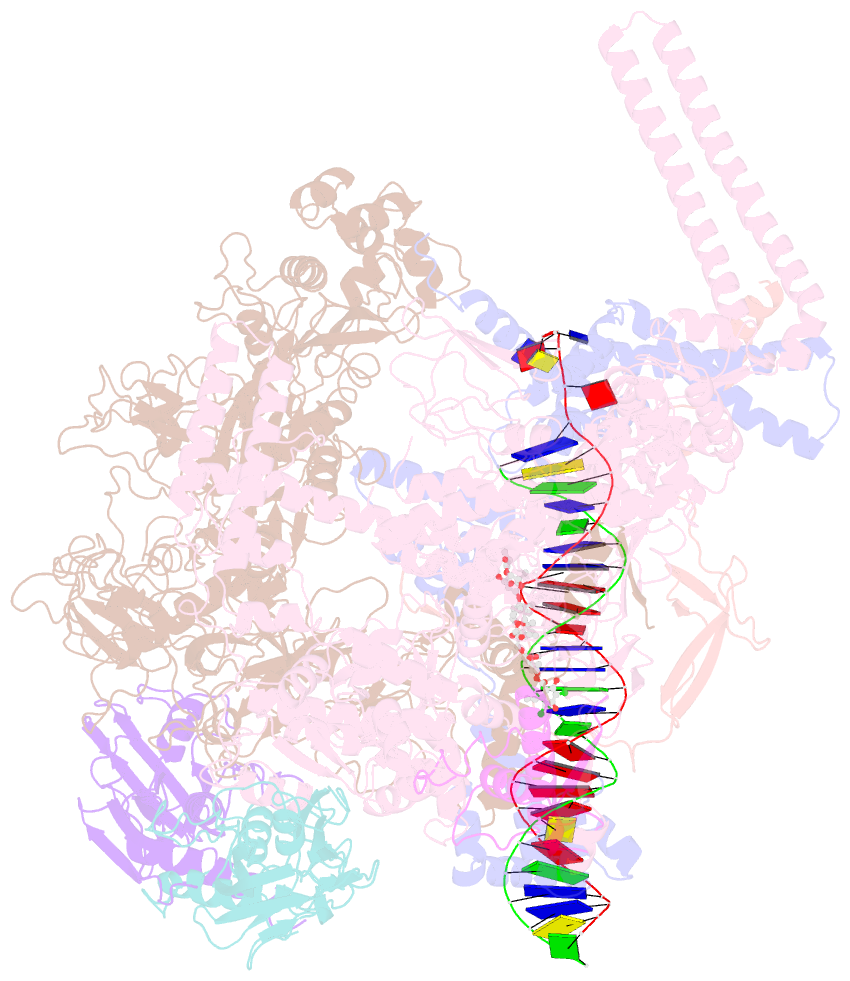
Summary information and primary citation
- PDB-id
- 6bzo; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA-antibiotic
- Method
- cryo-EM (3.38 Å)
- Summary
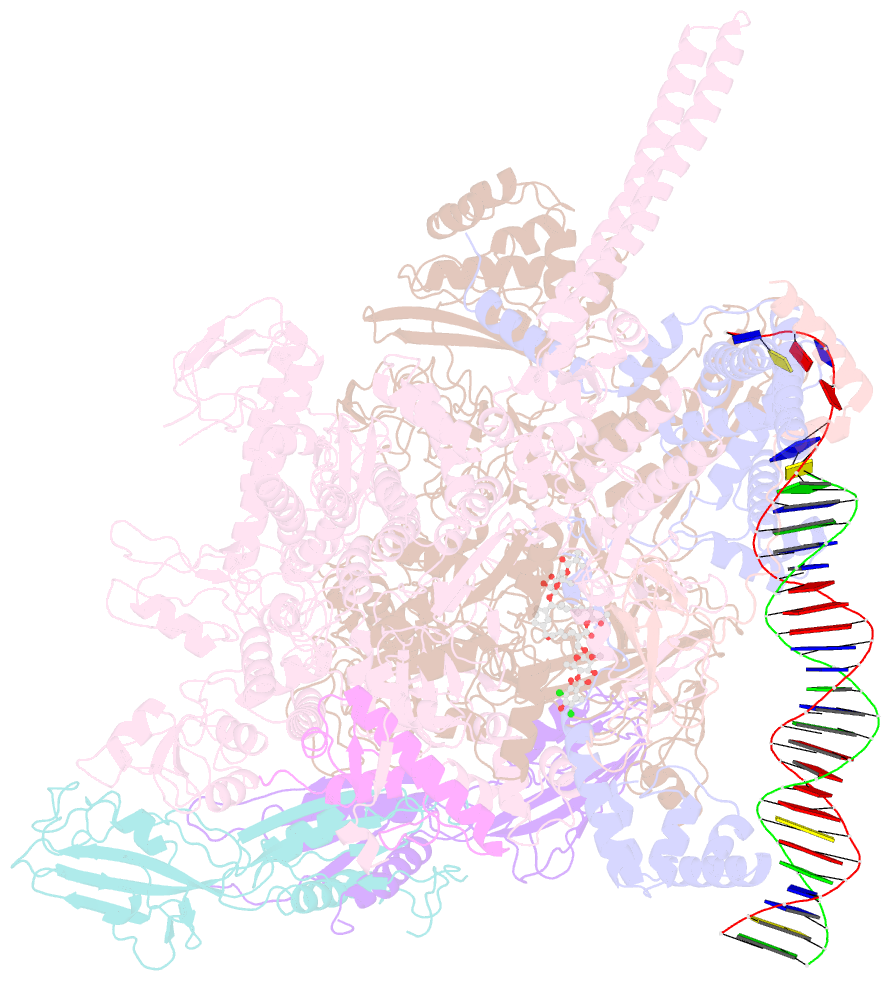
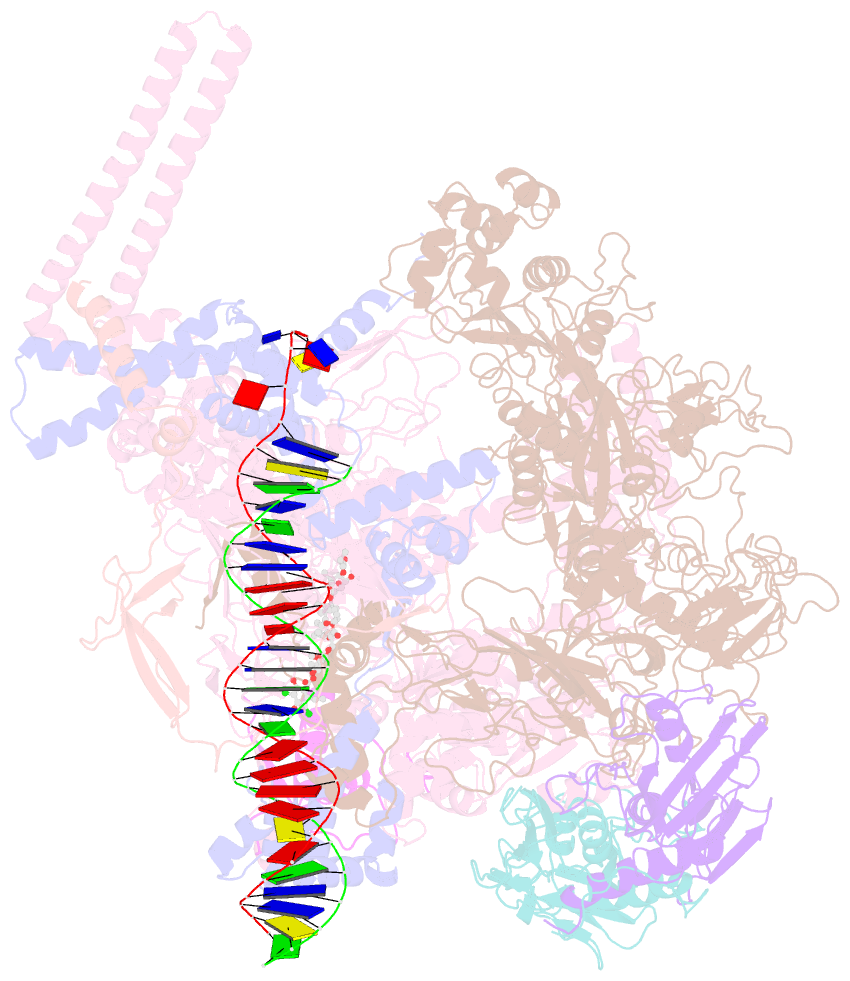
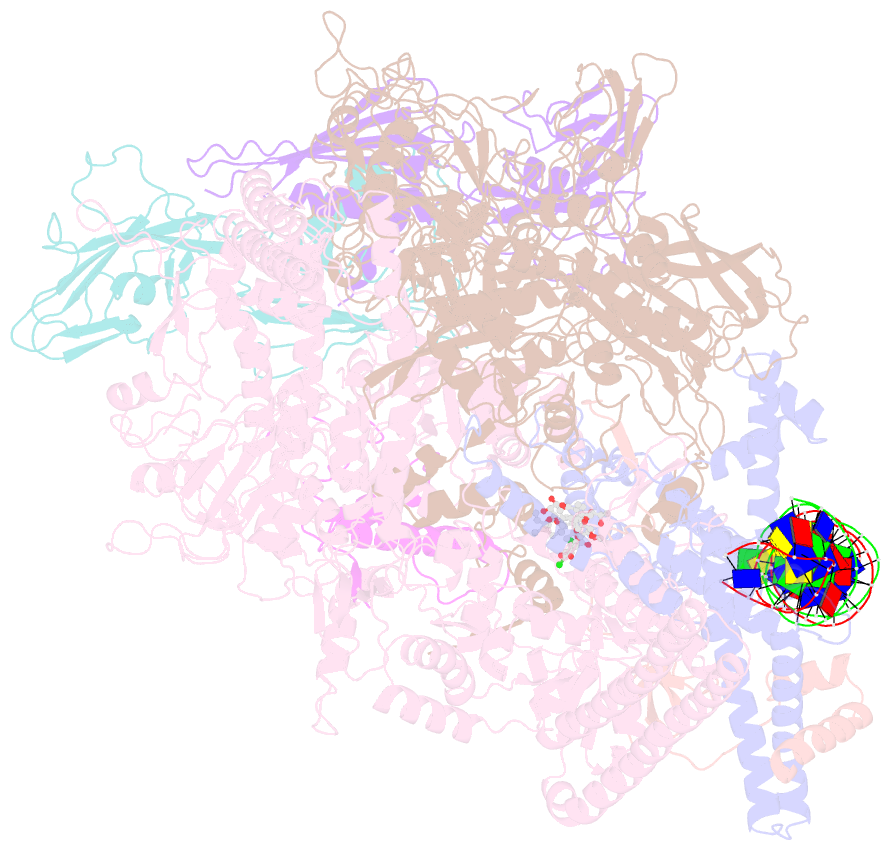
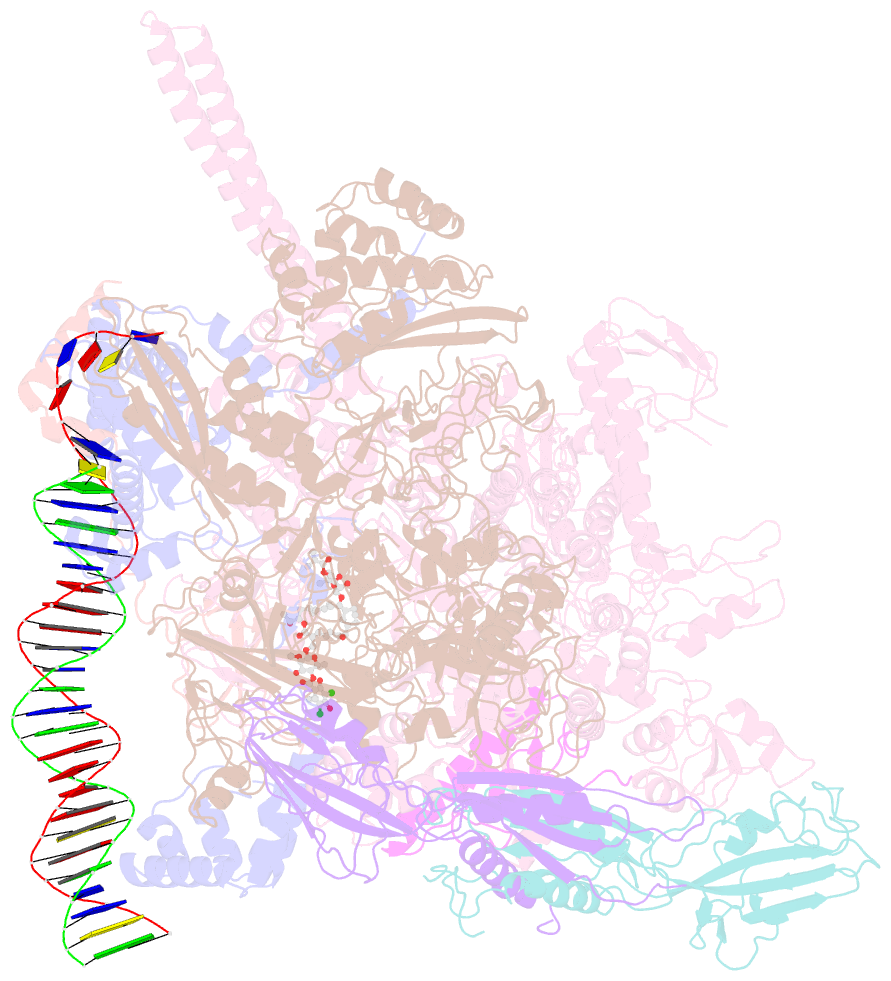
- Mtb rnap holo-rbpa-fidaxomicin-upstream fork DNA
- Reference
- Boyaci H, Chen J, Lilic M, Palka M, Mooney RA, Landick R, Darst SA, Campbell EA (2018): "Fidaxomicin jamsMycobacterium tuberculosisRNA polymerase motions needed for initiation via RbpA contacts." Elife, 7. doi: 10.7554/eLife.34823.
- Abstract
- Fidaxomicin (Fdx) is an antimicrobial RNA polymerase (RNAP) inhibitor highly effective against Mycobacterium tuberculosis RNAP in vitro, but clinical use of Fdx is limited to treating Clostridium difficile intestinal infections due to poor absorption. To identify the structural determinants of Fdx binding to RNAP, we determined the 3.4 Å cryo-electron microscopy structure of a complete M. tuberculosis RNAP holoenzyme in complex with Fdx. We find that the actinobacteria general transcription factor RbpA contacts fidaxomycin, explaining its strong effect on M. tuberculosis. Additional structures define conformational states of M. tuberculosis RNAP between the free apo-holoenzyme and the promoter-engaged open complex ready for transcription. The results establish that Fdx acts like a doorstop to jam the enzyme in an open state, preventing the motions necessary to secure promoter DNA in the active site. Our results provide a structural platform to guide development of anti-tuberculosis antimicrobials based on the Fdx binding pocket.