Summary information and primary citation
- PDB-id
- 6c0w; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- structural protein-DNA
- Method
- cryo-EM (4.0 Å)
- Summary
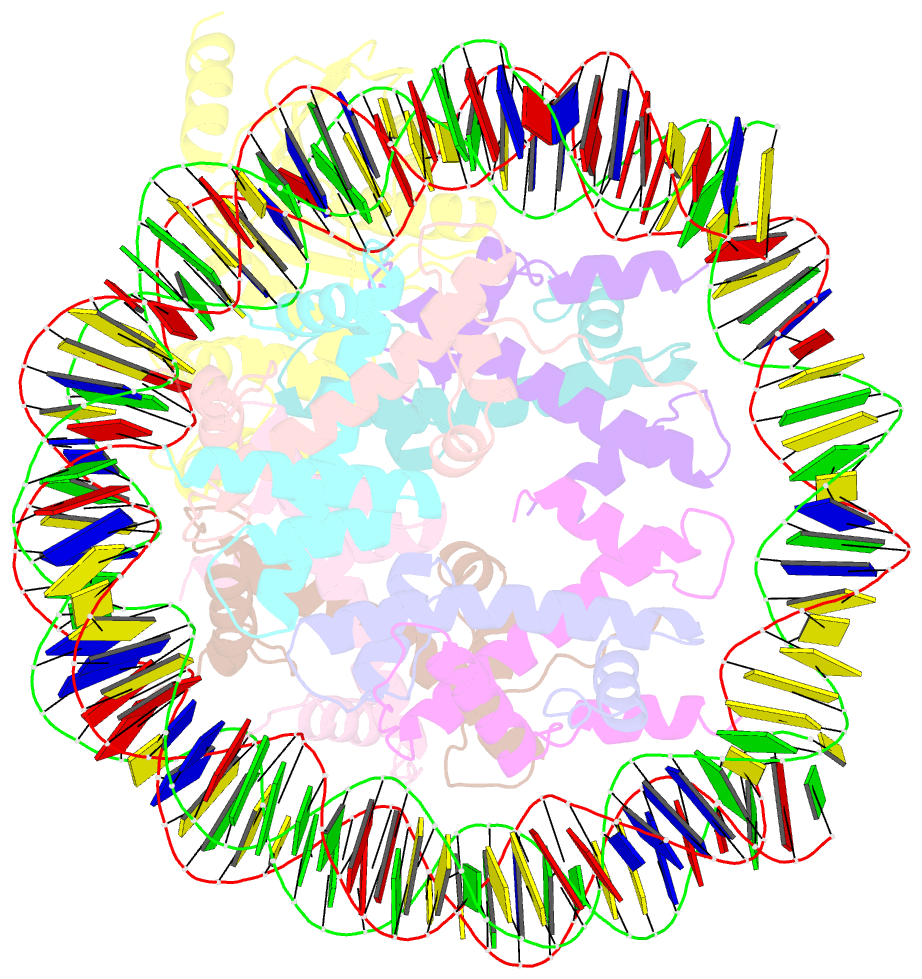
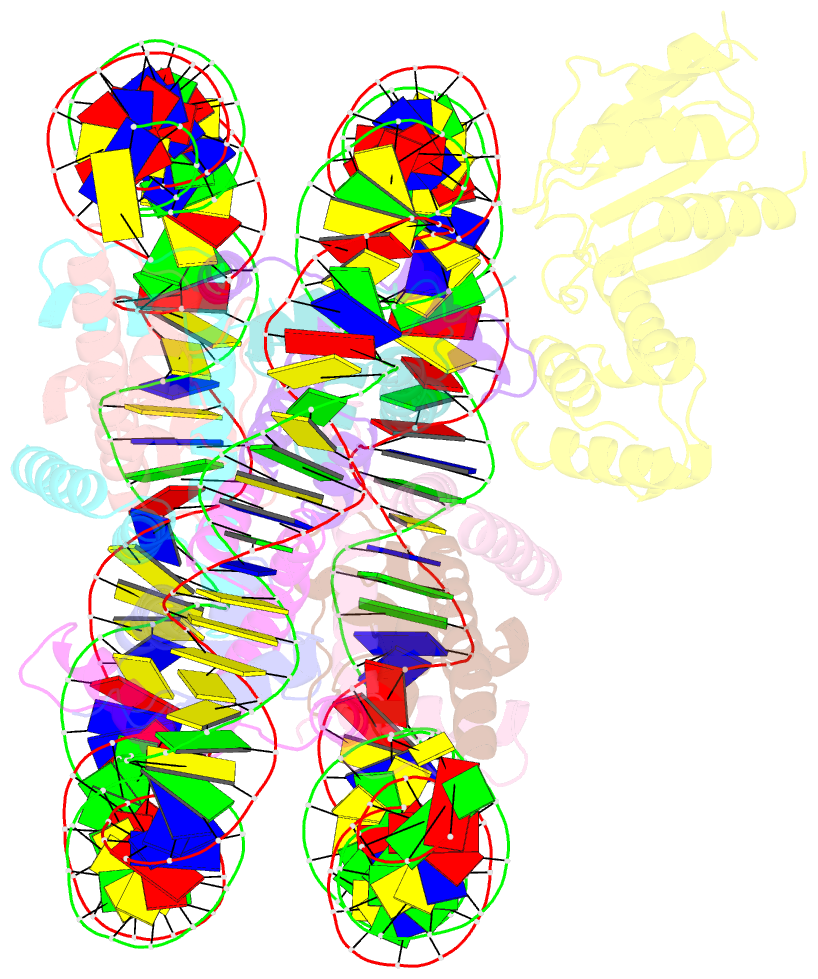
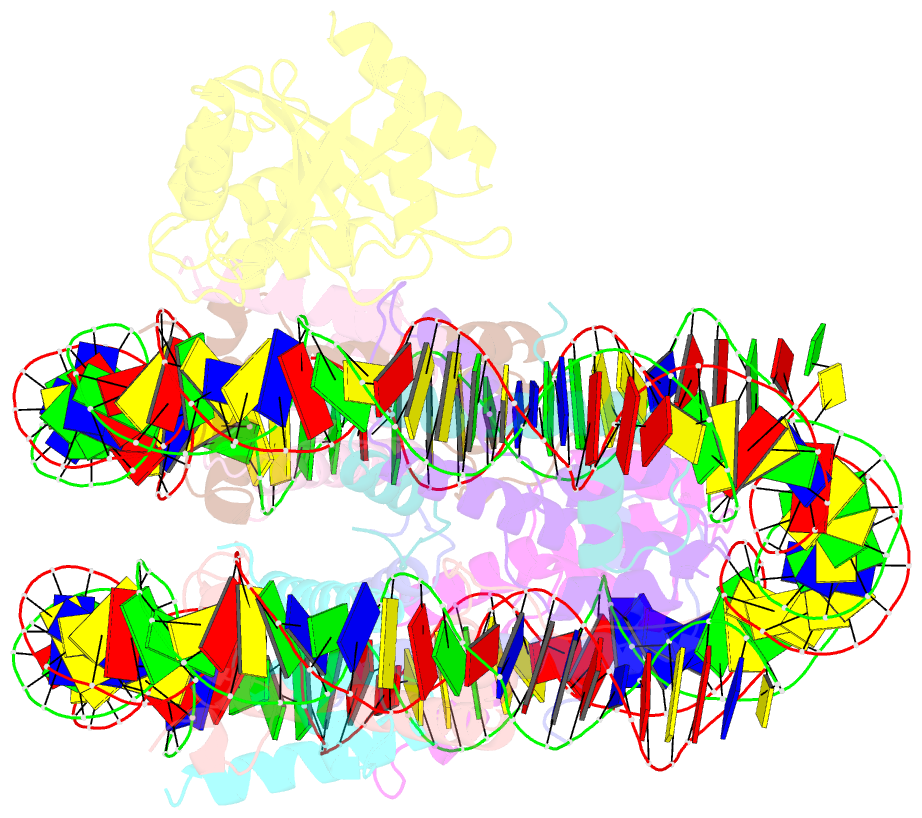
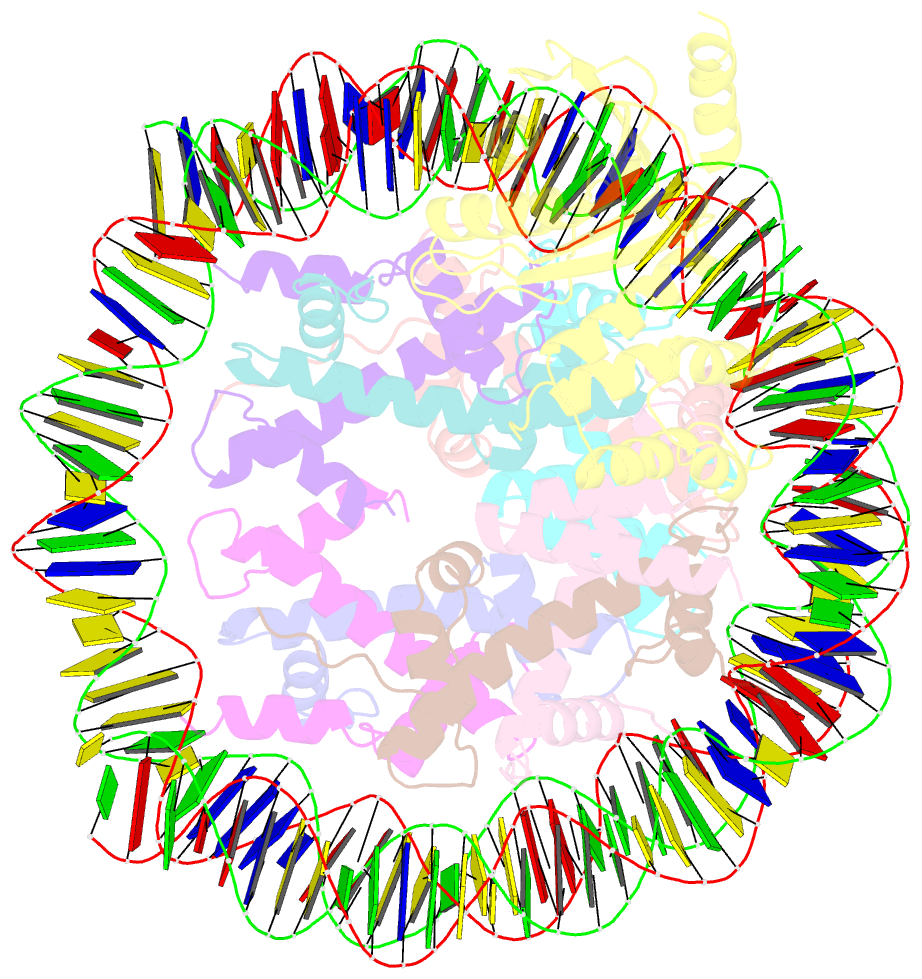
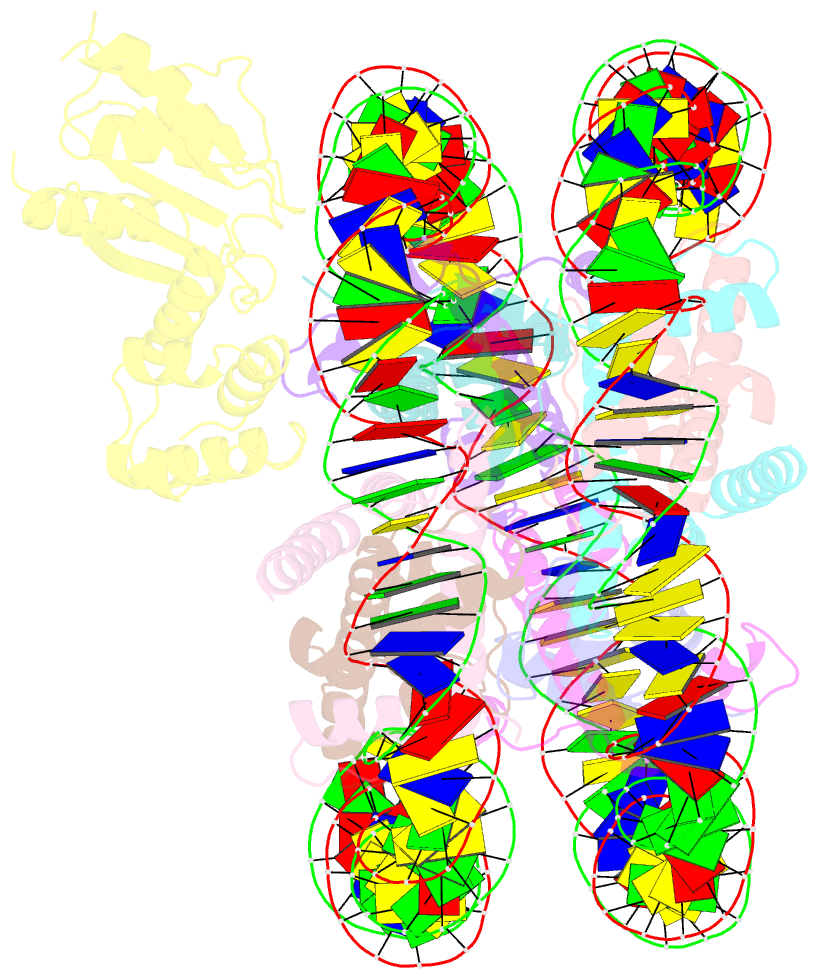
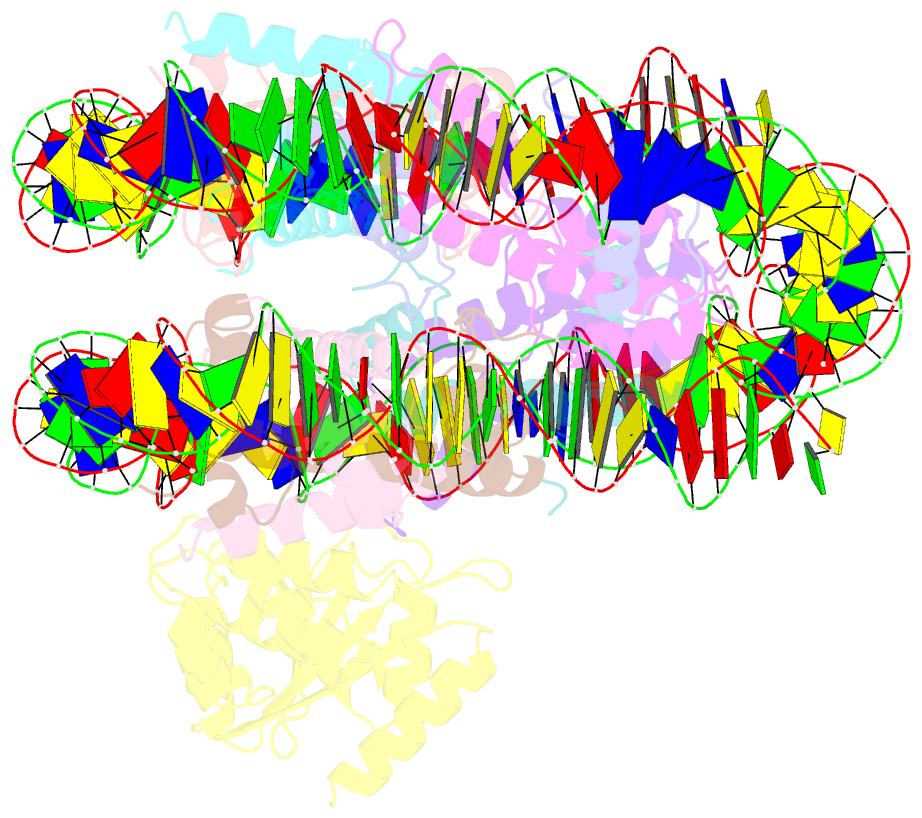
- cryo-EM structure of human kinetochore protein cenp-n with the centromeric nucleosome containing cenp-a
- Reference
- Pentakota S, Zhou K, Smith C, Maffini S, Petrovic A, Morgan GP, Weir JR, Vetter IR, Musacchio A, Luger K (2017): "Decoding the centromeric nucleosome through CENP-N." Elife, 6. doi: 10.7554/eLife.33442.
- Abstract
- Centromere protein (CENP) A, a histone H3 variant, is a key epigenetic determinant of chromosome domains known as centromeres. Centromeres nucleate kinetochores, multi-subunit complexes that capture spindle microtubules to promote chromosome segregation during mitosis. Two kinetochore proteins, CENP-C and CENP-N, recognize CENP-A in the context of a rare CENP-A nucleosome. Here, we reveal the structural basis for the exquisite selectivity of CENP-N for centromeres. CENP-N uses charge and space complementarity to decode the L1 loop that is unique to CENP-A. It also engages in extensive interactions with a 15-base pair segment of the distorted nucleosomal DNA double helix, in a position predicted to exclude chromatin remodelling enzymes. Besides CENP-A, stable centromere recruitment of CENP-N requires a coincident interaction with a newly identified binding motif on nucleosome-bound CENP-C. Collectively, our studies clarify how CENP-N and CENP-C decode and stabilize the non-canonical CENP-A nucleosome to enforce epigenetic centromere specification and kinetochore assembly.