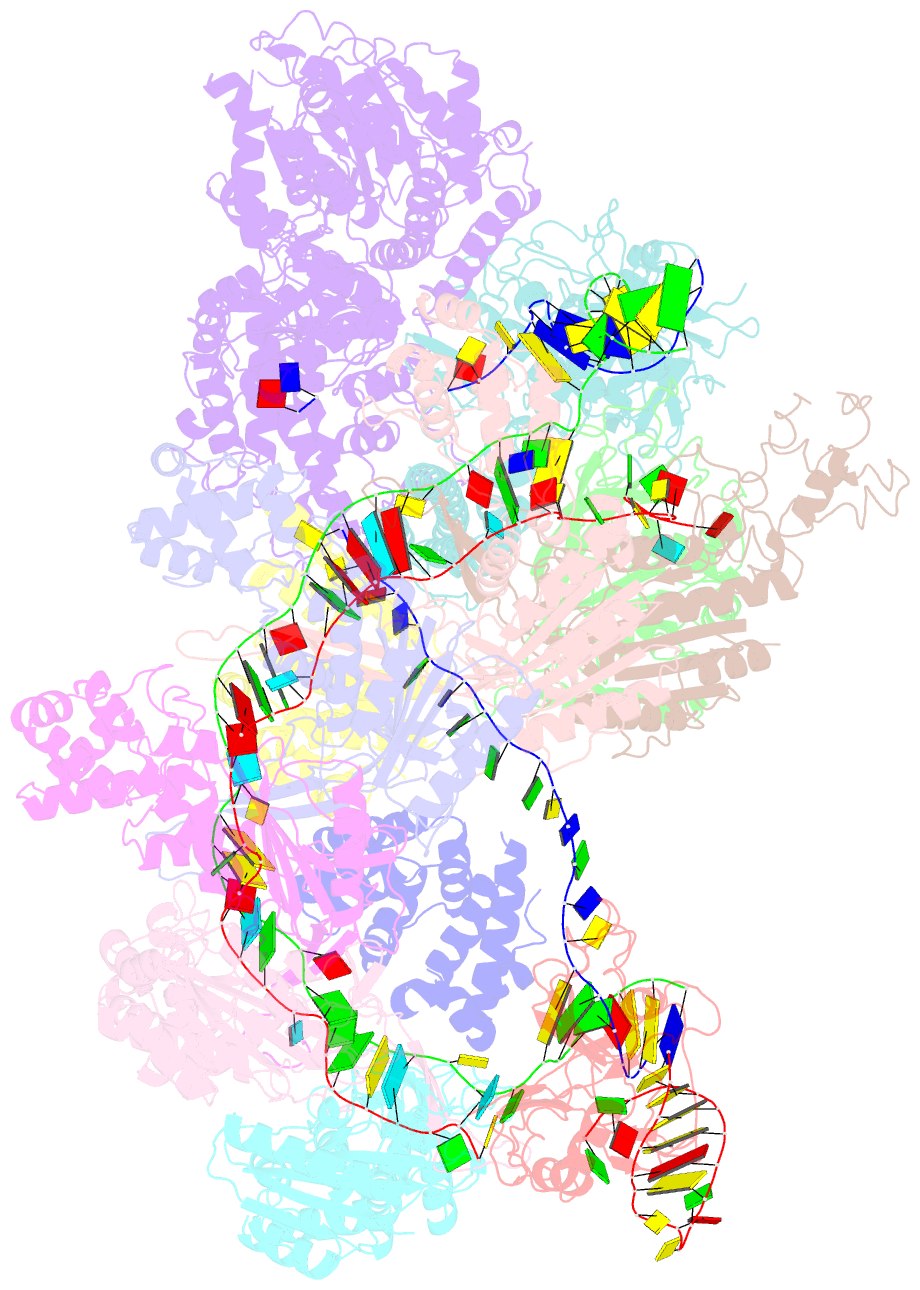
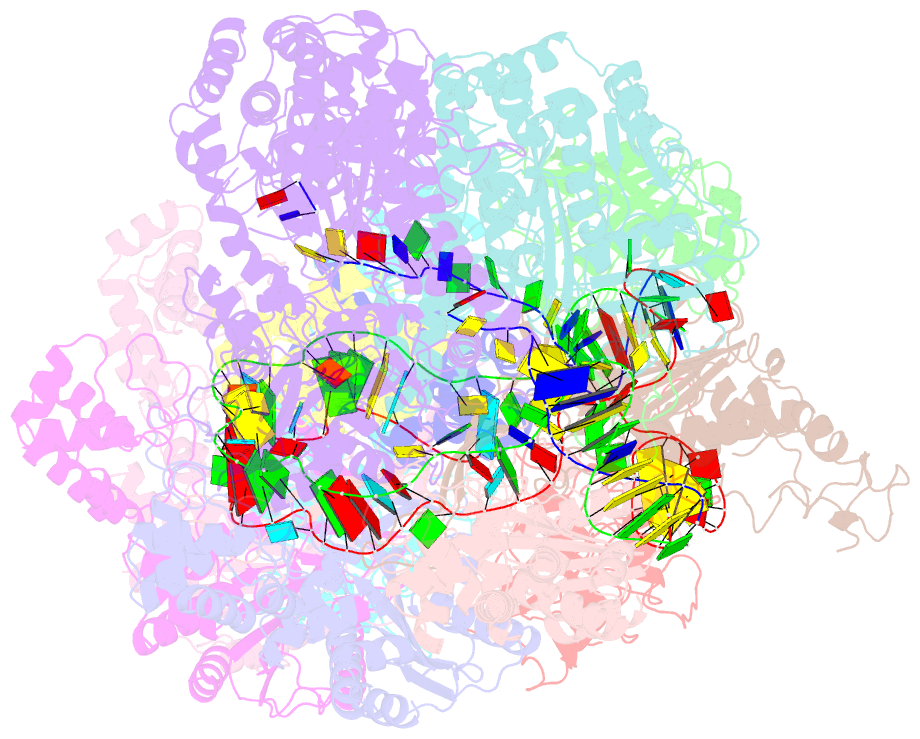
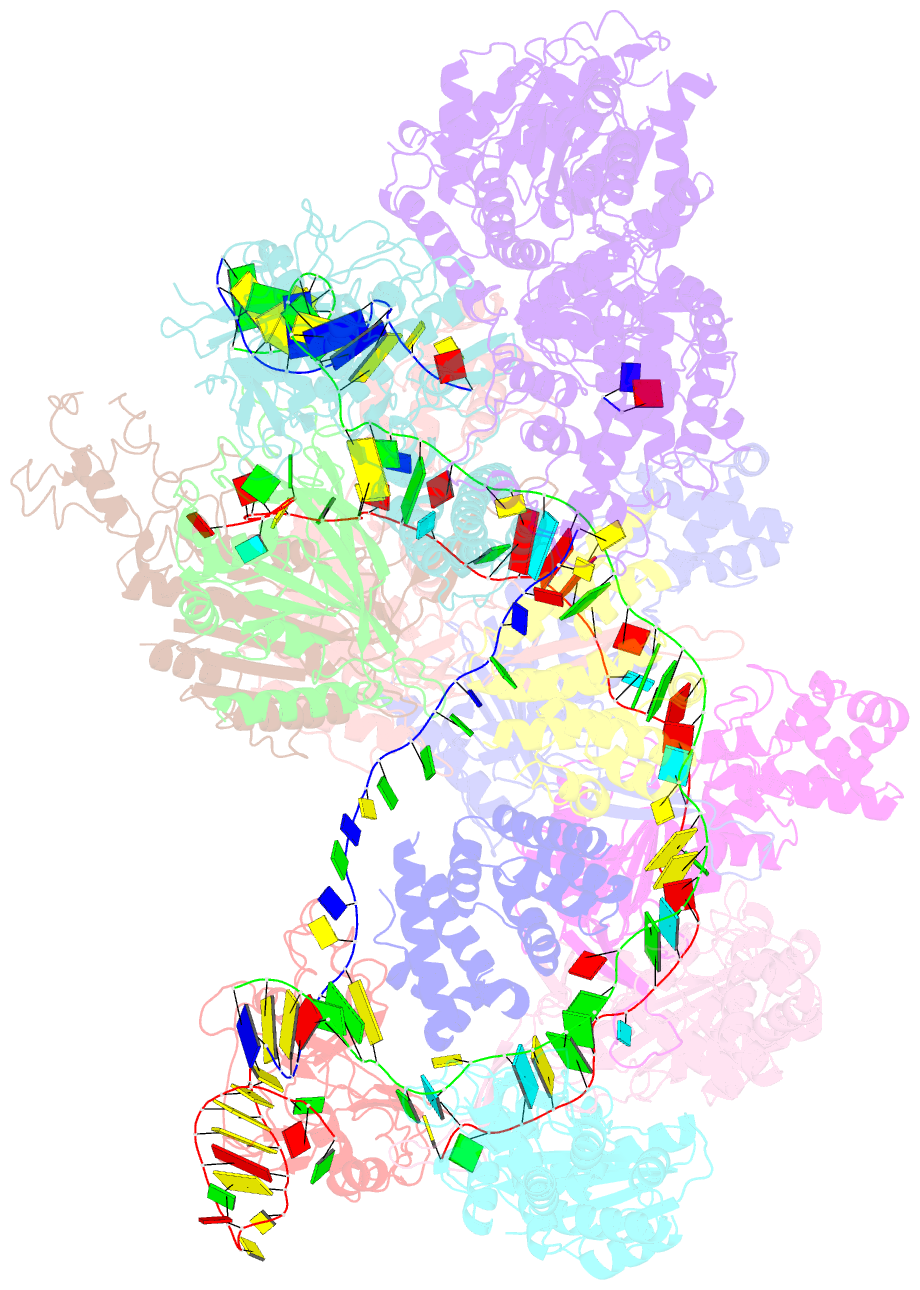
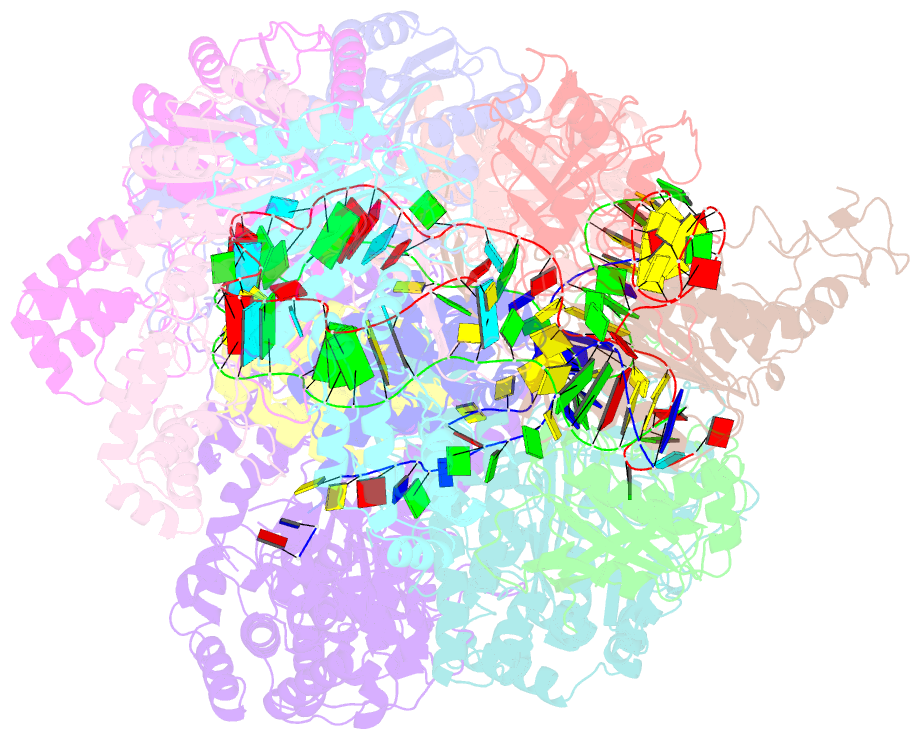
Summary information and primary citation
- PDB-id
- 6c66; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA-RNA
- Method
- cryo-EM (3.66 Å)
- Summary
- Crispr RNA-guided surveillance complex, pre-nicking
- Reference
- Xiao Y, Luo M, Dolan AE, Liao M, Ke A (2018): "Structure basis for RNA-guided DNA degradation by Cascade and Cas3." Science, 361. doi: 10.1126/science.aat0839.
- Abstract
- Type I CRISPR-Cas system features a sequential target-searching and degradation process on double-stranded DNA by the RNA-guided Cascade (CRISPR associated complex for antiviral defense) complex and the nuclease-helicase fusion enzyme Cas3, respectively. Here, we present a 3.7-angstrom-resolution cryo-electron microscopy (cryo-EM) structure of the Type I-E Cascade/R-loop/Cas3 complex, poised to initiate DNA degradation. Cas3 distinguishes Cascade conformations and only captures the R-loop-forming Cascade, to avoid cleaving partially complementary targets. Its nuclease domain recruits the nontarget strand (NTS) DNA at a bulged region for the nicking of single-stranded DNA. An additional 4.7-angstrom-resolution cryo-EM structure captures the postnicking state, in which the severed NTS retracts to the helicase entrance, to be threaded for adenosine 5'-triphosphate-dependent processive degradation. These snapshots form the basis for understanding RNA-guided DNA degradation in Type I-E CRISPR-Cas systems.