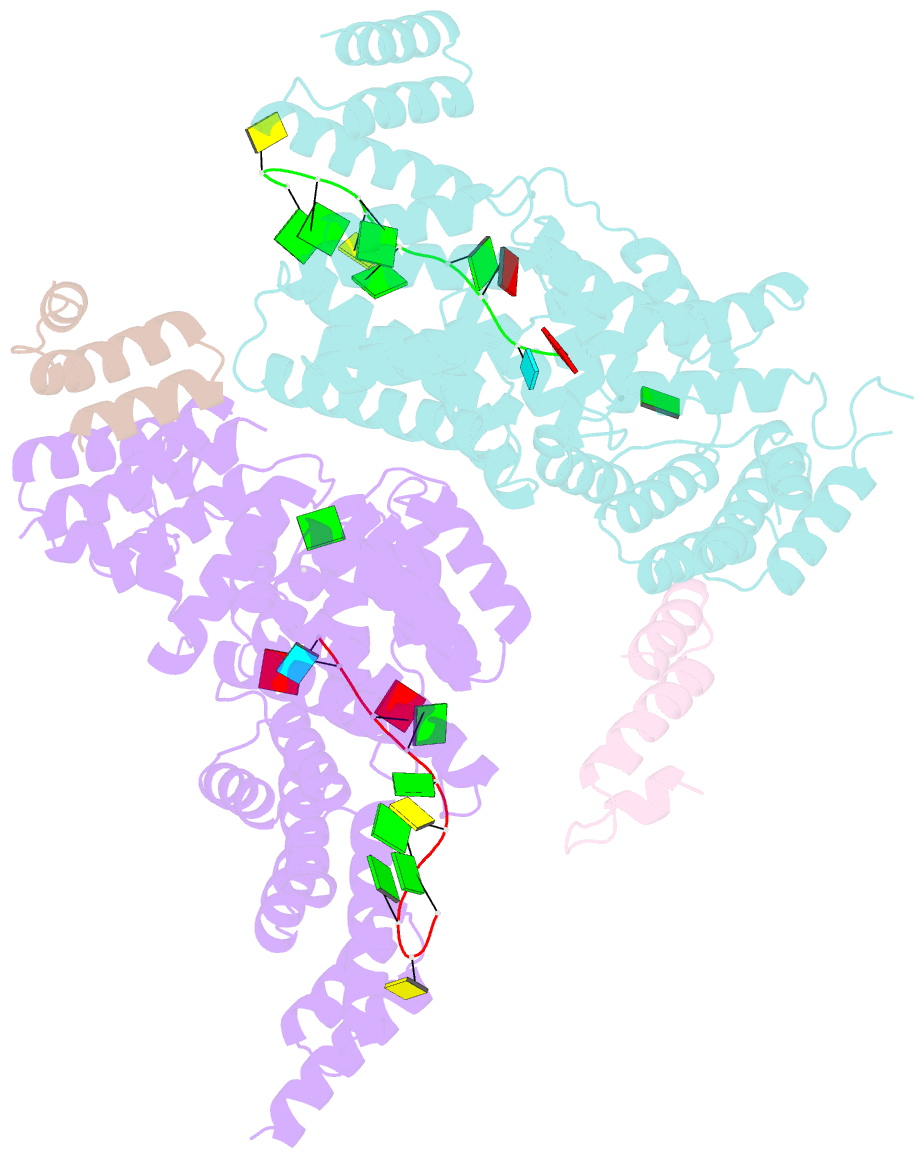
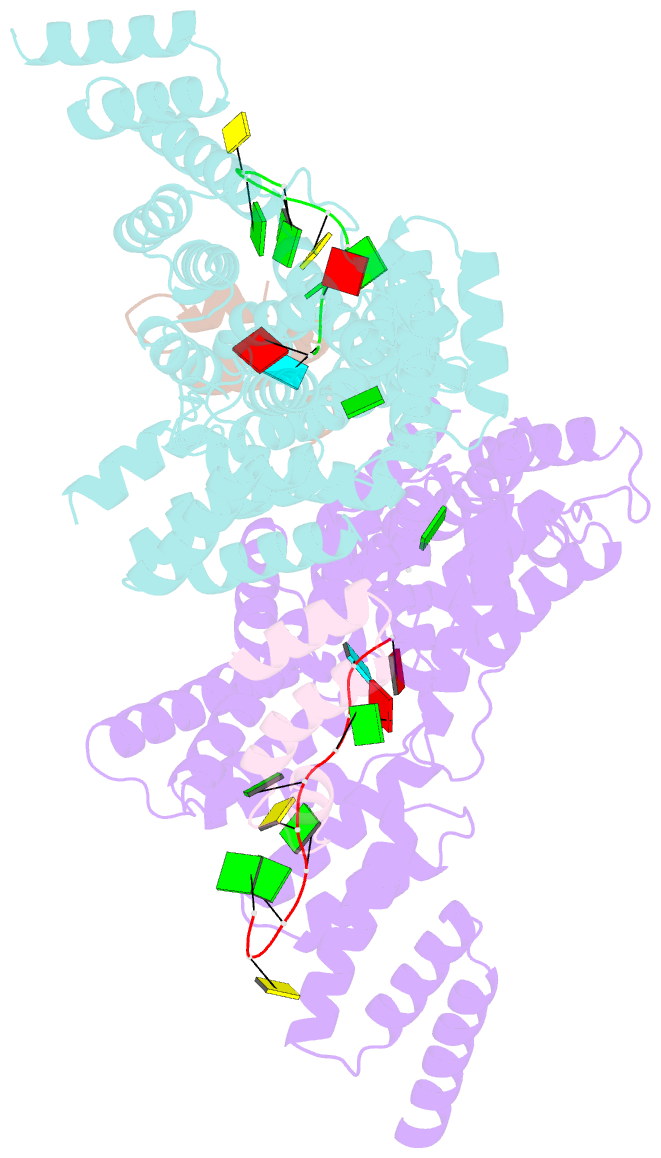
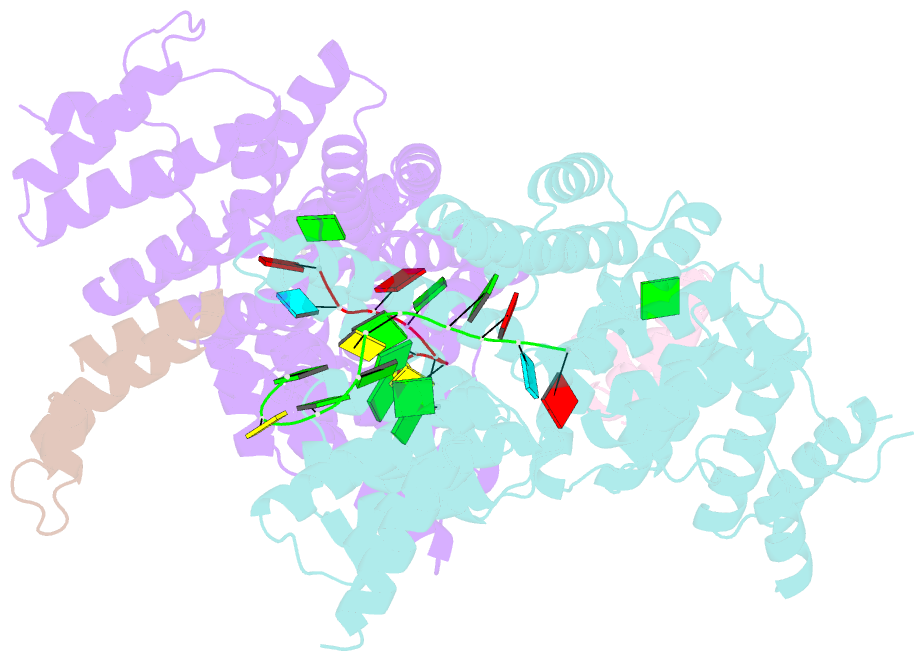
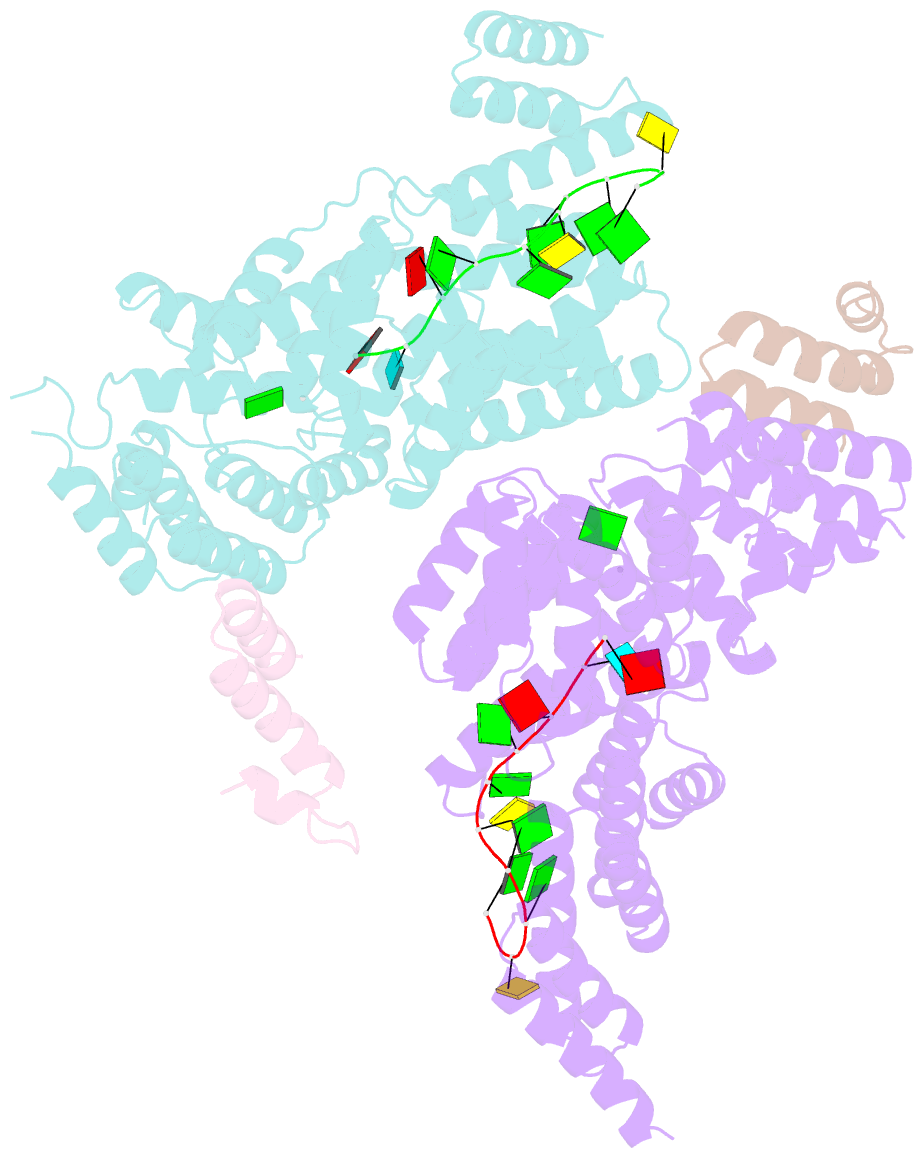
Summary information and primary citation
- PDB-id
- 6c6k; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- X-ray (2.54 Å)
- Summary
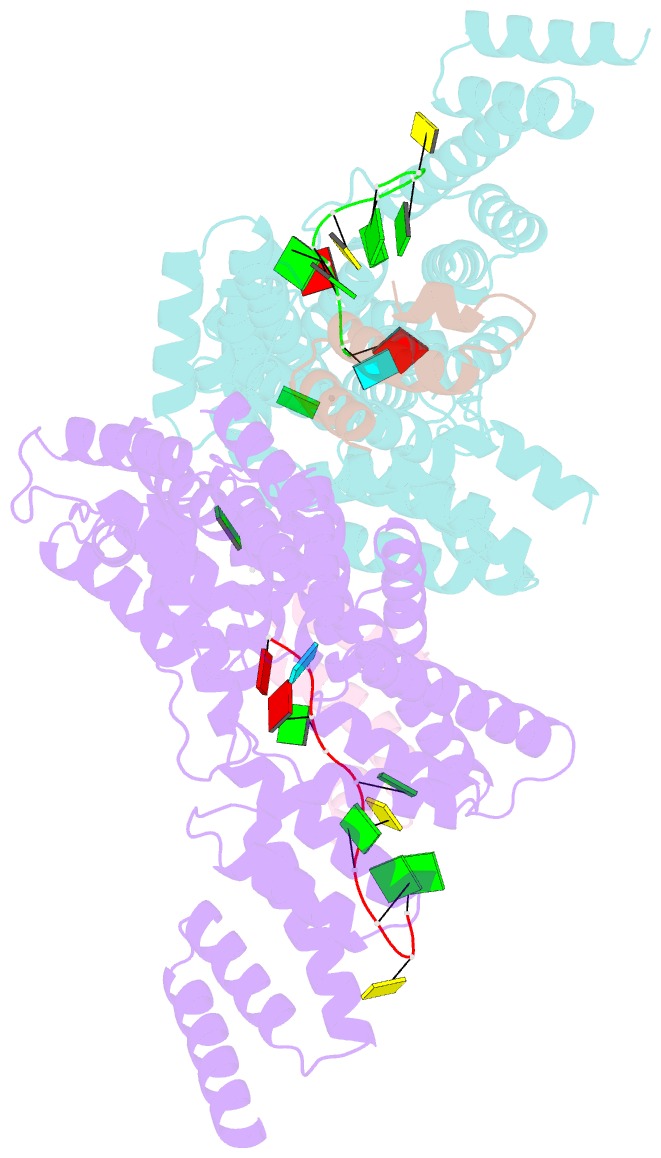
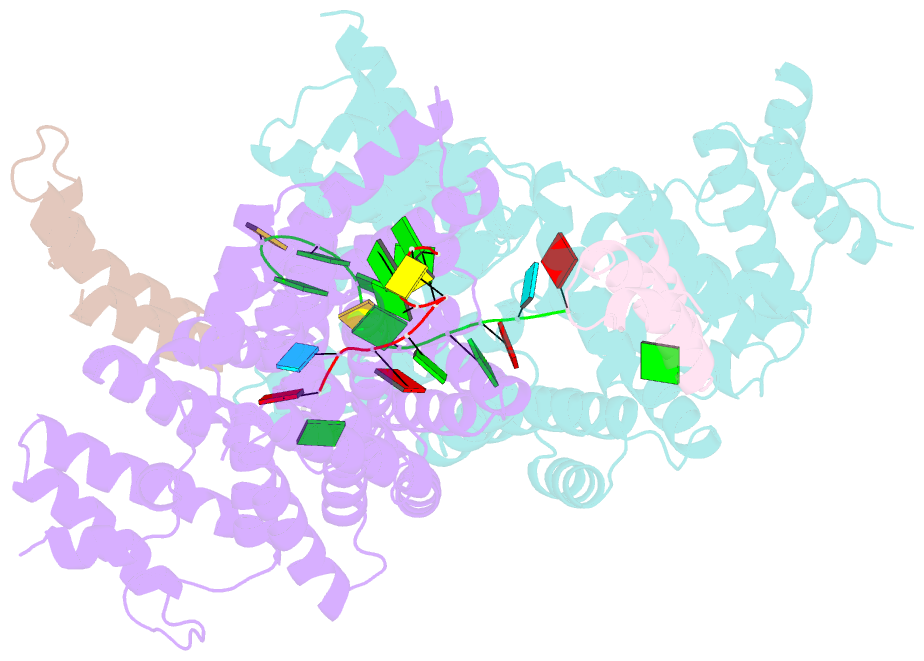
- Structural basis for preferential recognition of cap 0 RNA by a human ifit1-ifit3 protein complex
- Reference
- Johnson B, VanBlargan LA, Xu W, White JP, Shan C, Shi PY, Zhang R, Adhikari J, Gross ML, Leung DW, Diamond MS, Amarasinghe GK (2018): "Human IFIT3 Modulates IFIT1 RNA Binding Specificity and Protein Stability." Immunity, 48, 487-499.e5. doi: 10.1016/j.immuni.2018.01.014.
- Abstract
- Although interferon-induced proteins with tetratricopeptide repeats (IFIT proteins) inhibit infection of many viruses by recognizing their RNA, the regulatory mechanisms involved remain unclear. Here we report a crystal structure of cap 0 (m7GpppN) RNA bound to human IFIT1 in complex with the C-terminal domain of human IFIT3. Structural, biochemical, and genetic studies suggest that IFIT3 binding to IFIT1 has dual regulatory functions: (1) extending the half-life of IFIT1 and thereby increasing its steady-state amounts in cells; and (2) allosterically regulating the IFIT1 RNA-binding channel, thereby enhancing the specificity of recognition for cap 0 but not cap 1 (m7GpppNm) or 5'-ppp RNA. Mouse Ifit3 lacks this key C-terminal domain and does not bind mouse Ifit1. The IFIT3 interaction with IFIT1 is important for restricting infection of viruses lacking 2'-O methylation in their RNA cap structures. Our experiments establish differences in the regulation of IFIT1 orthologs and define targets for modulation of human IFIT protein activity.