Summary information and primary citation
- PDB-id
- 6c6s; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA-RNA
- Method
- cryo-EM (3.7 Å)
- Summary
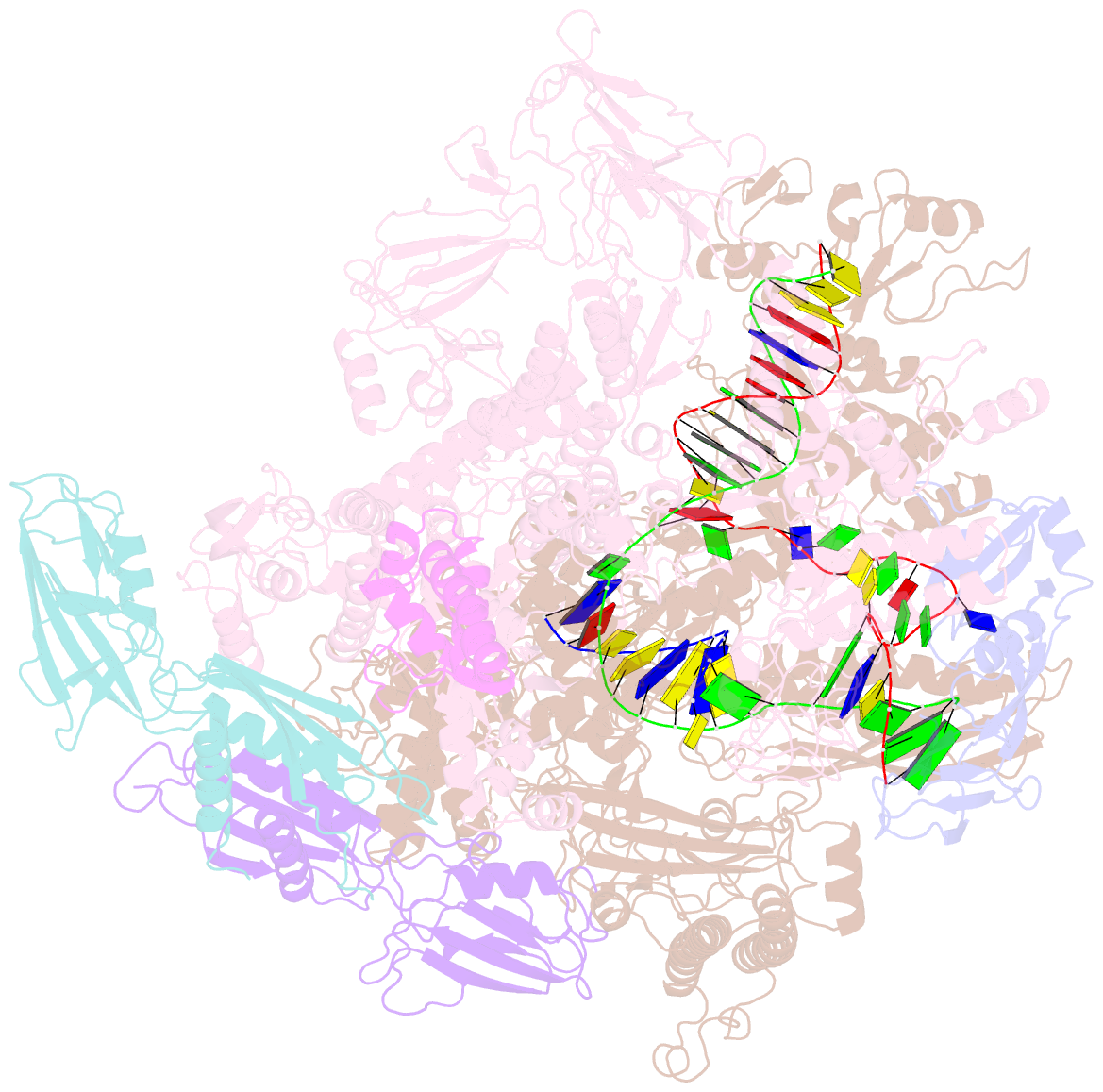
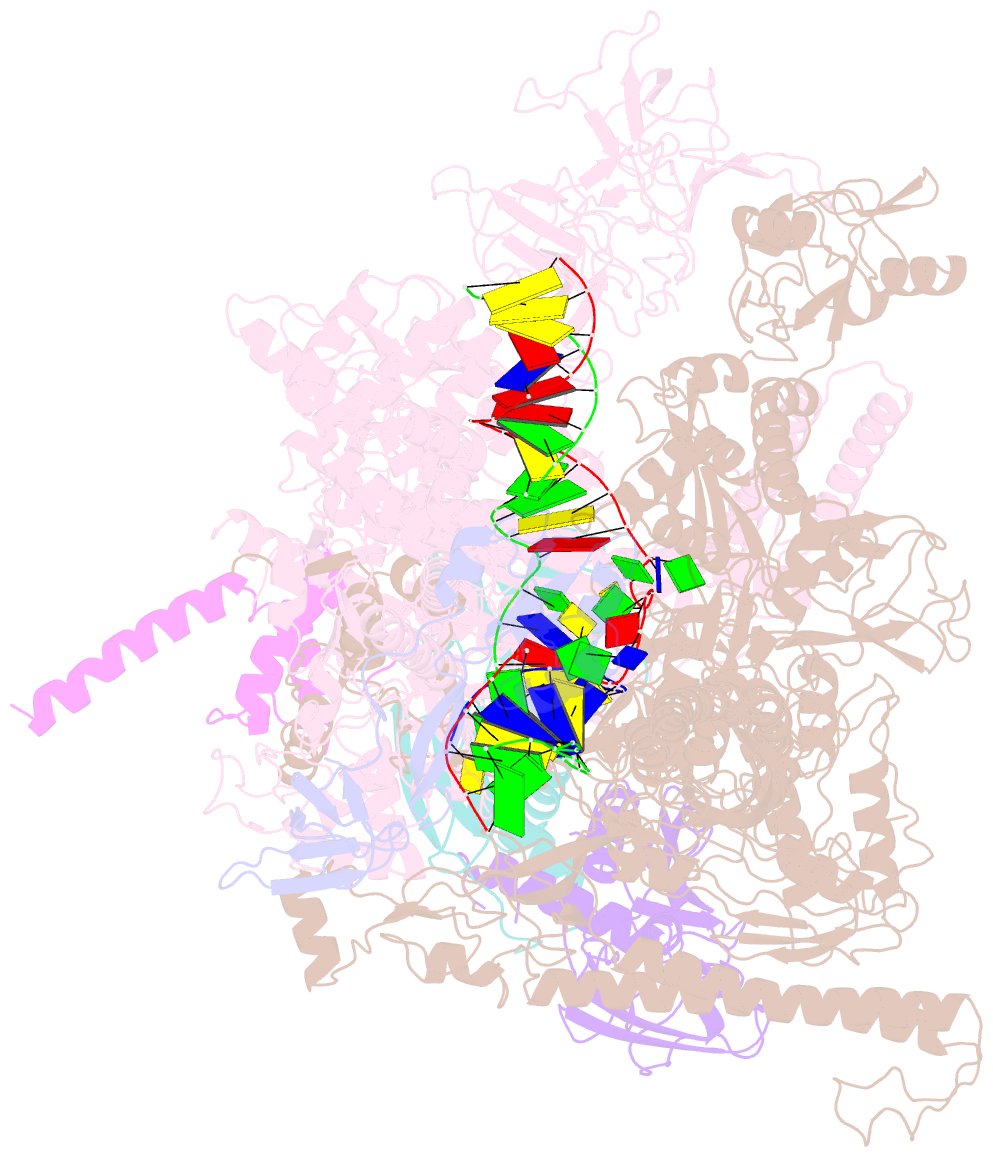
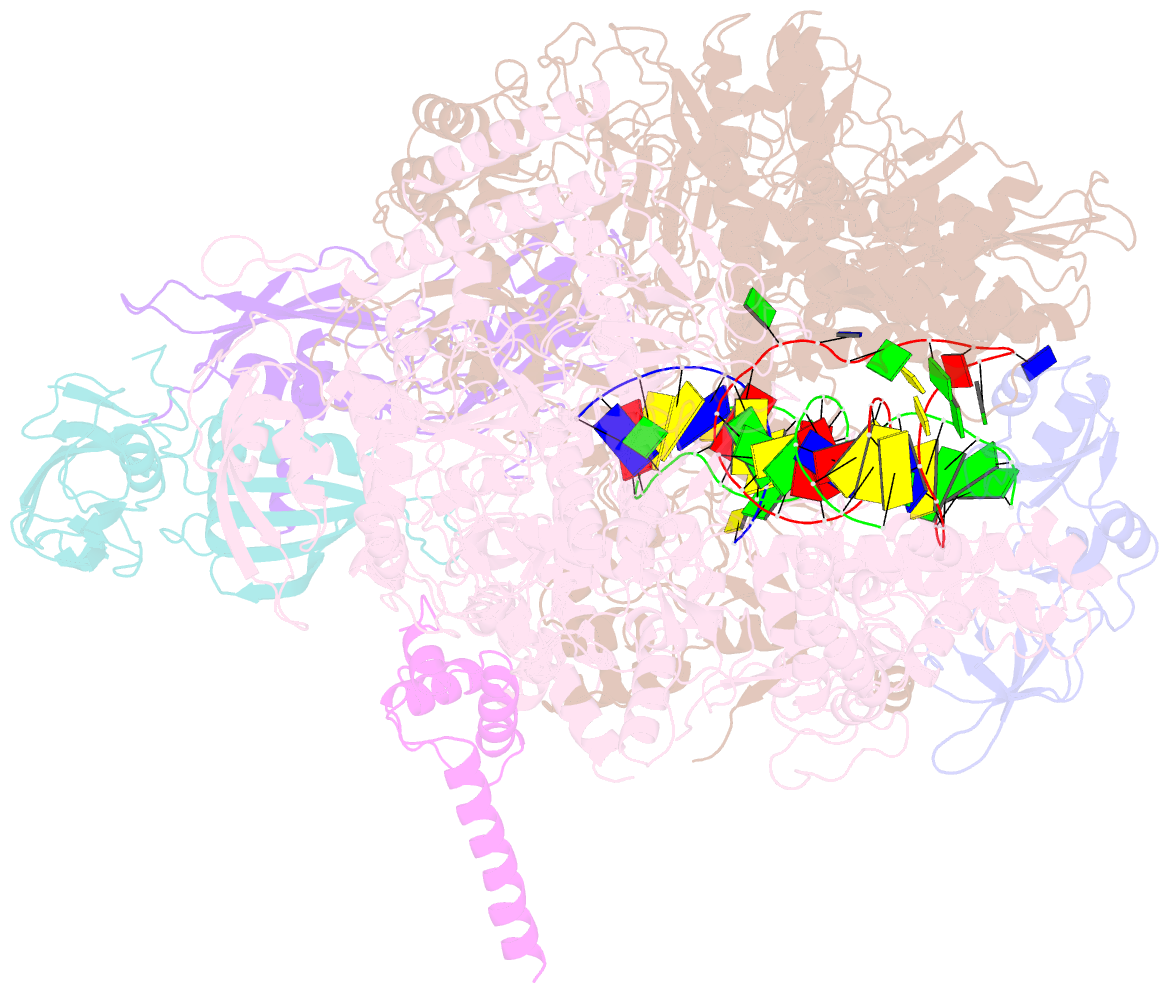
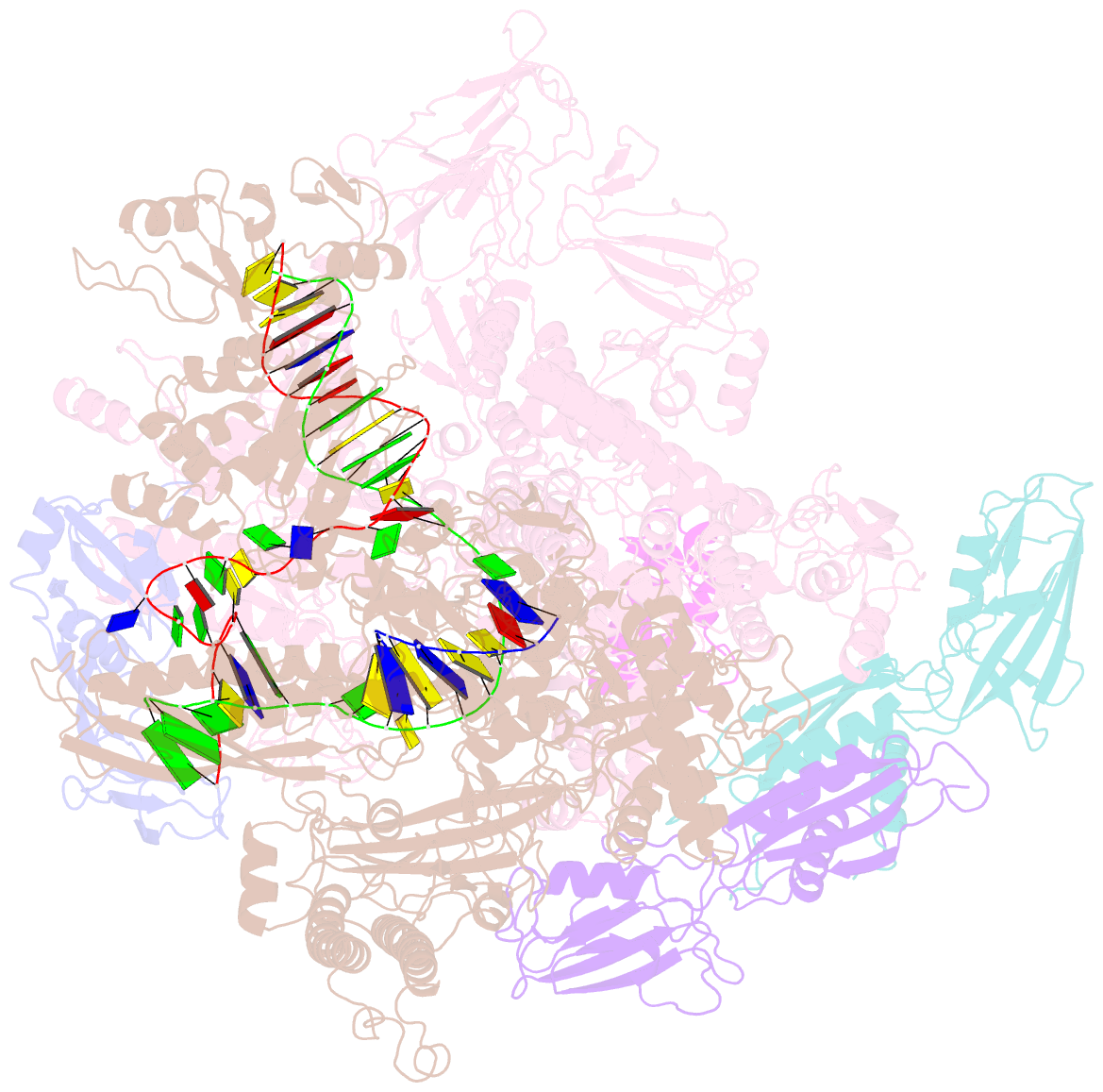
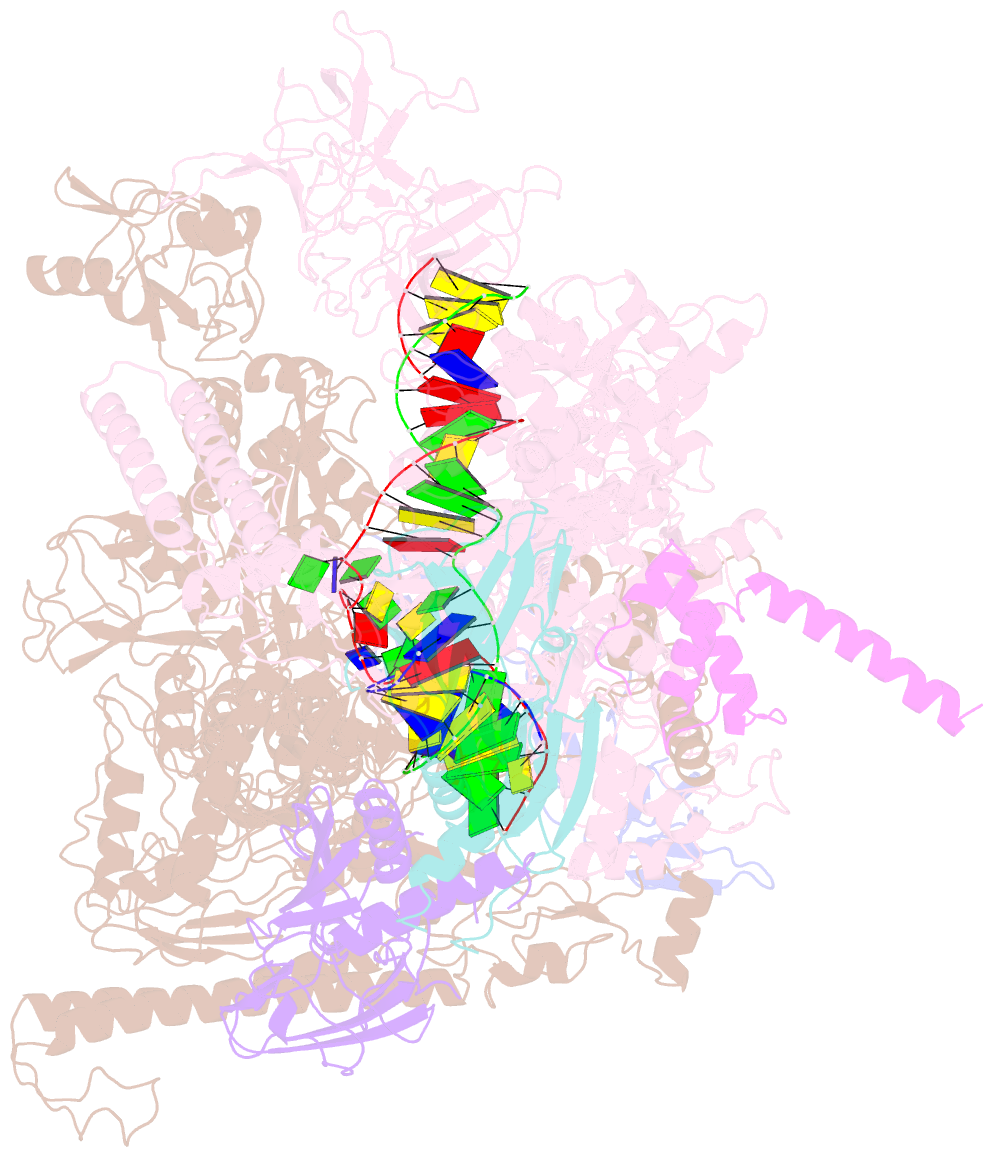
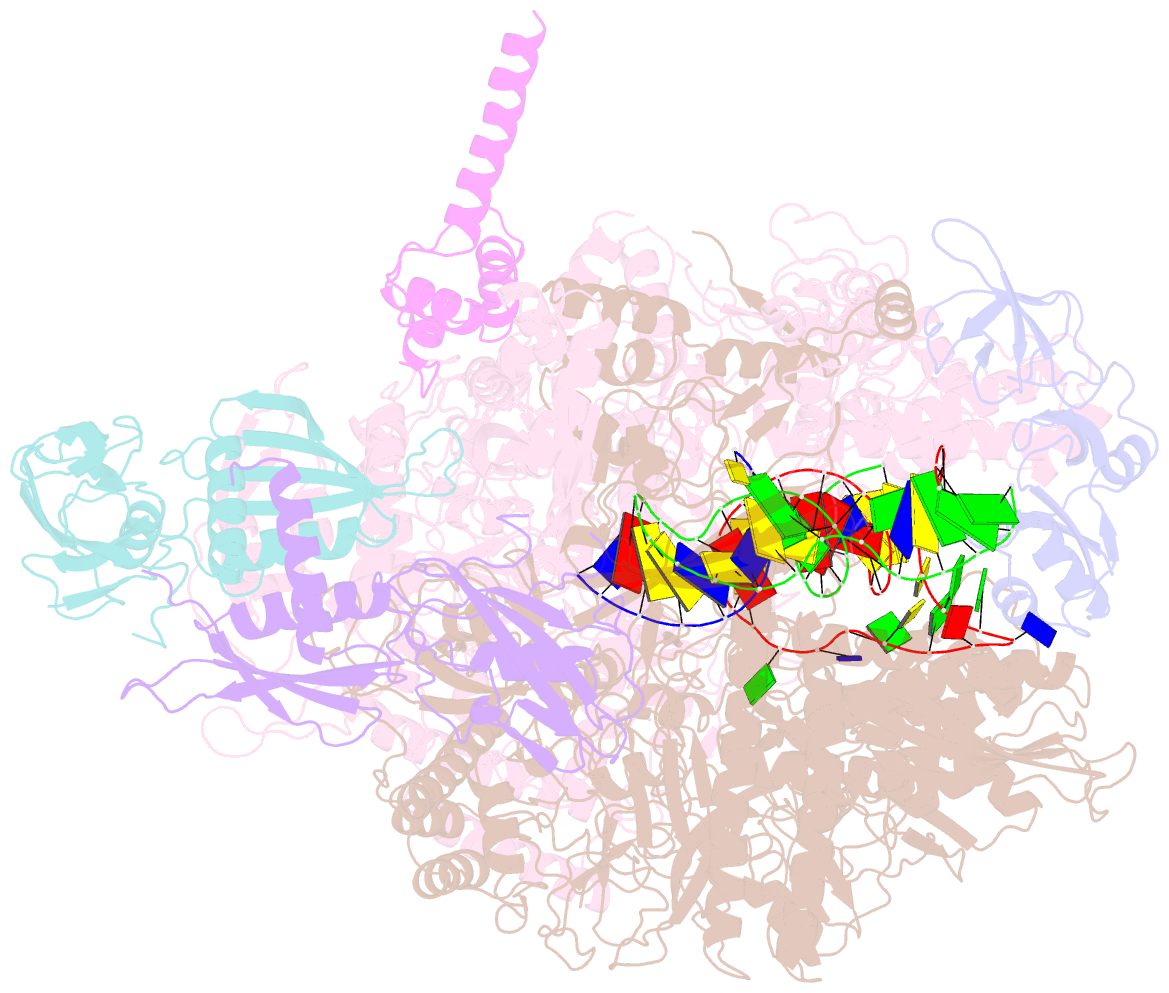
- Cryoem structure of e.coli RNA polymerase elongation complex bound with rfah
- Reference
- Kang JY, Mooney RA, Nedialkov Y, Saba J, Mishanina TV, Artsimovitch I, Landick R, Darst SA (2018): "Structural Basis for Transcript Elongation Control by NusG Family Universal Regulators." Cell, 173, 1650-1662.e14. doi: 10.1016/j.cell.2018.05.017.
- Abstract
- NusG/RfaH/Spt5 transcription elongation factors are the only transcription regulators conserved across all life. Bacterial NusG regulates RNA polymerase (RNAP) elongation complexes (ECs) across most genes, enhancing elongation by suppressing RNAP backtracking and coordinating ρ-dependent termination and translation. The NusG paralog RfaH engages the EC only at operon polarity suppressor (ops) sites and suppresses both backtrack and hairpin-stabilized pausing. We used single-particle cryoelectron microscopy (cryo-EM) to determine structures of ECs at ops with NusG or RfaH. Both factors chaperone base-pairing of the upstream duplex DNA to suppress backtracking, explaining stimulation of elongation genome-wide. The RfaH-opsEC structure reveals how RfaH confers operon specificity through specific recognition of an ops hairpin in the single-stranded nontemplate DNA and tighter binding to the EC to exclude NusG. Tight EC binding by RfaH sterically blocks the swiveled RNAP conformation necessary for hairpin-stabilized pausing. The universal conservation of NusG/RfaH/Spt5 suggests that the molecular mechanisms uncovered here are widespread.