Summary information and primary citation
- PDB-id
- 6ca0; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- cryo-EM (5.75 Å)
- Summary
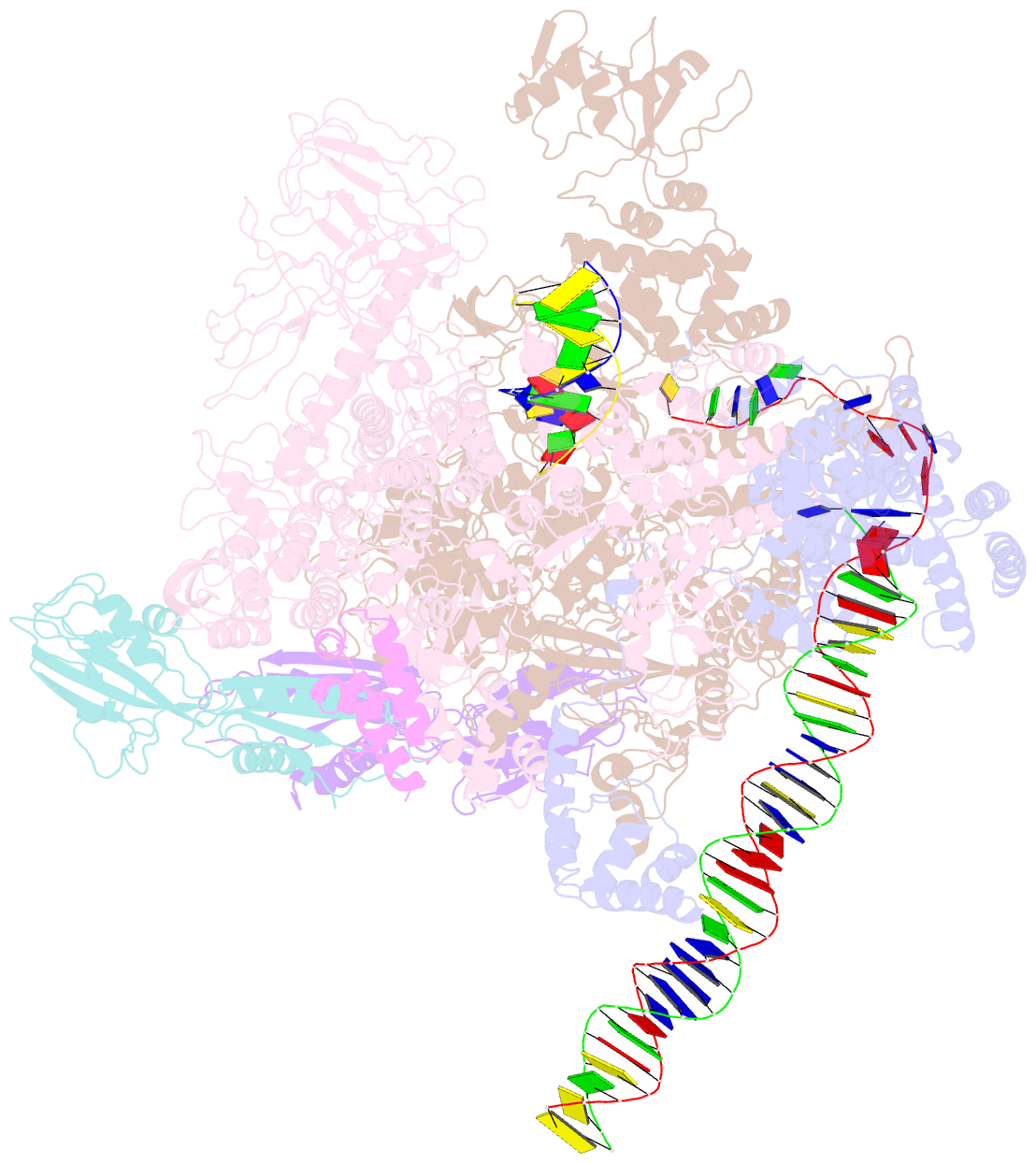
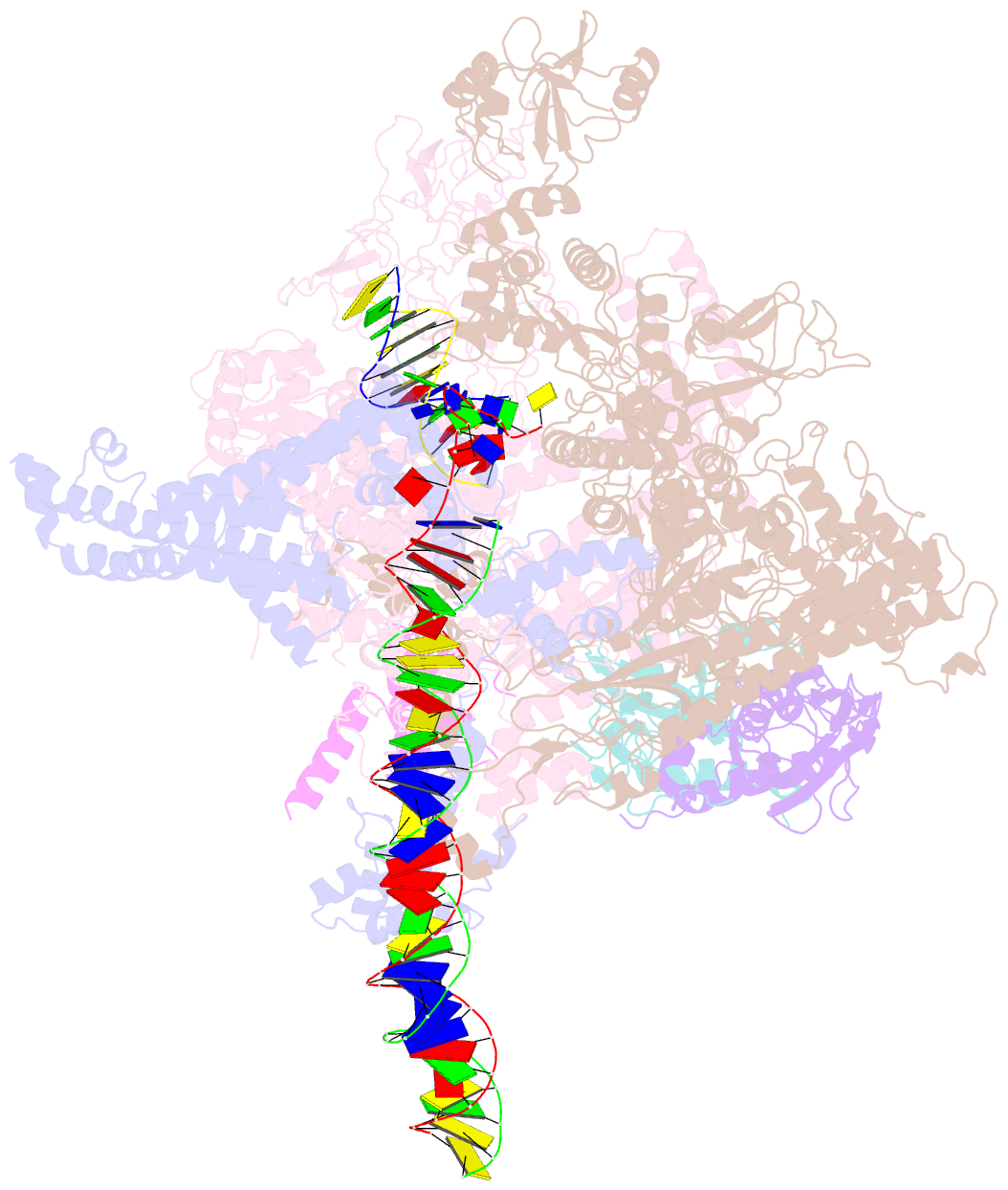
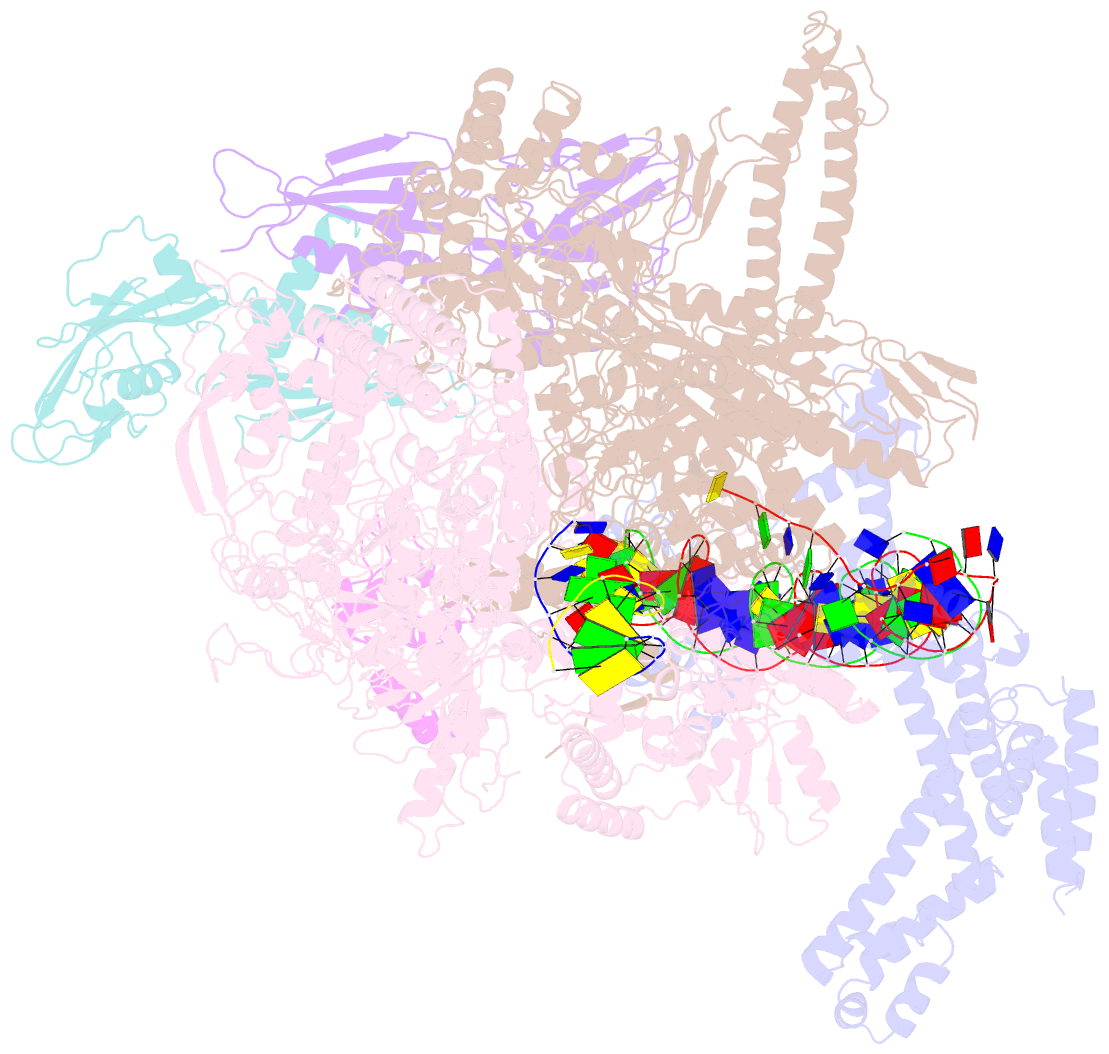
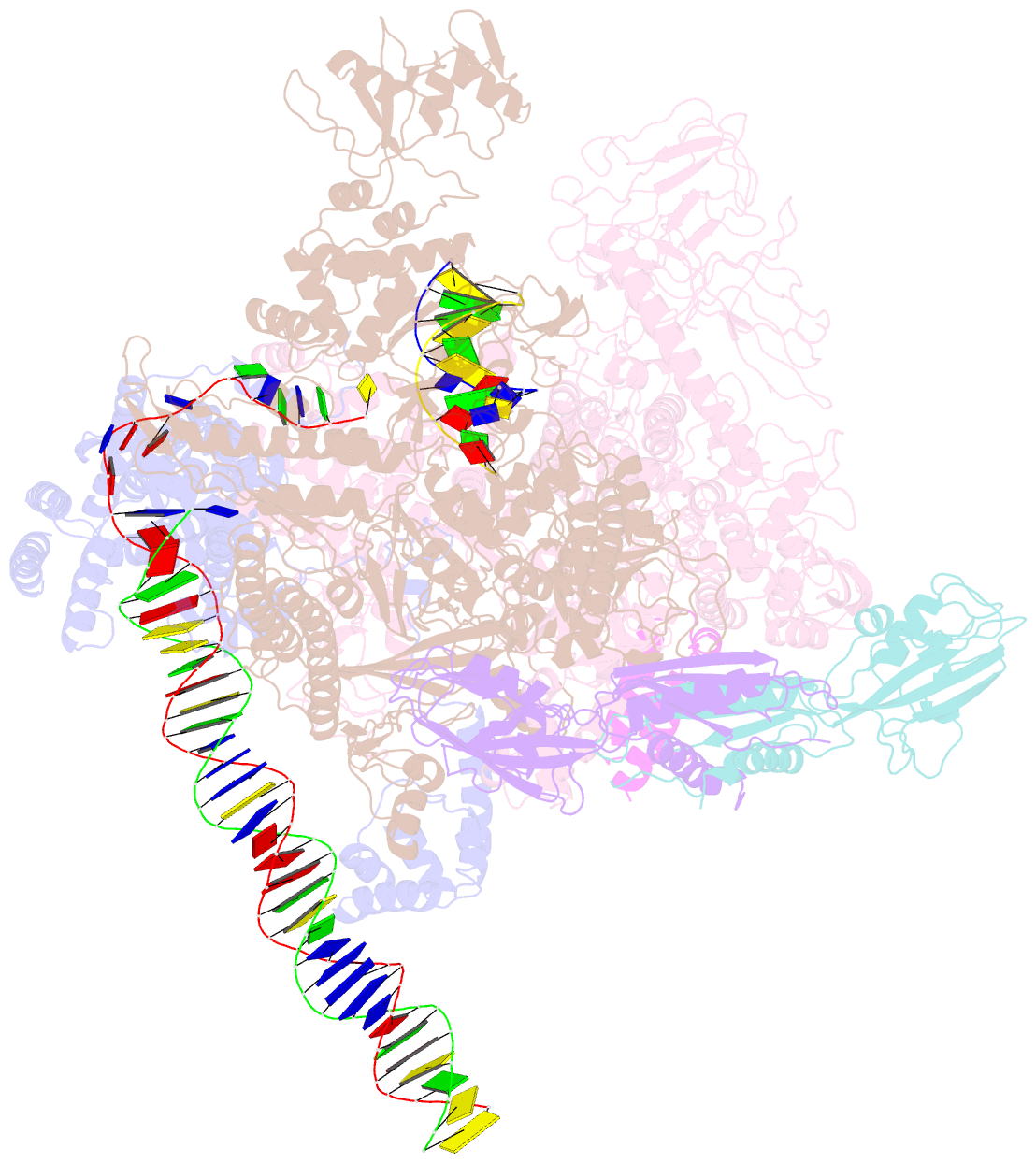
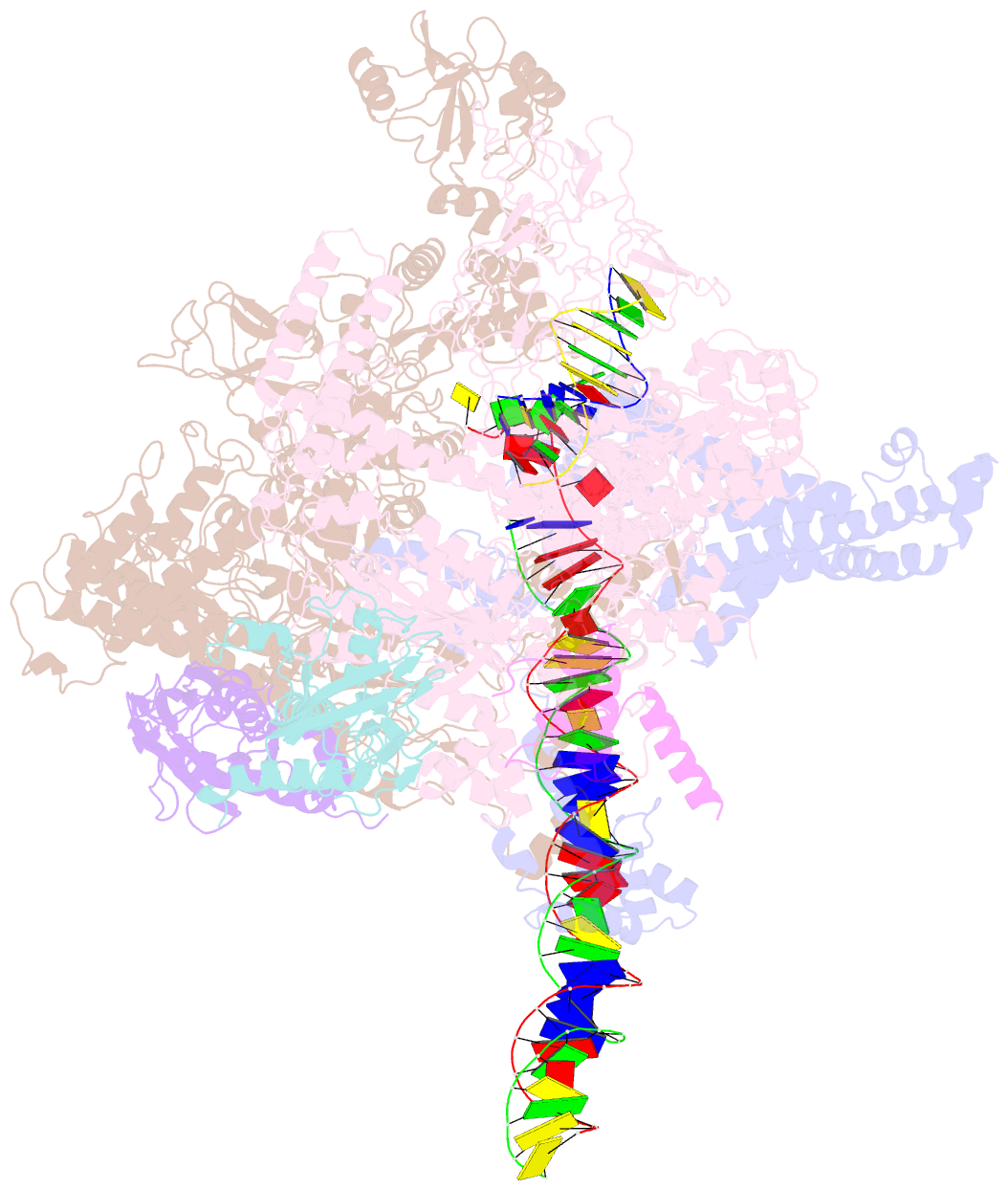
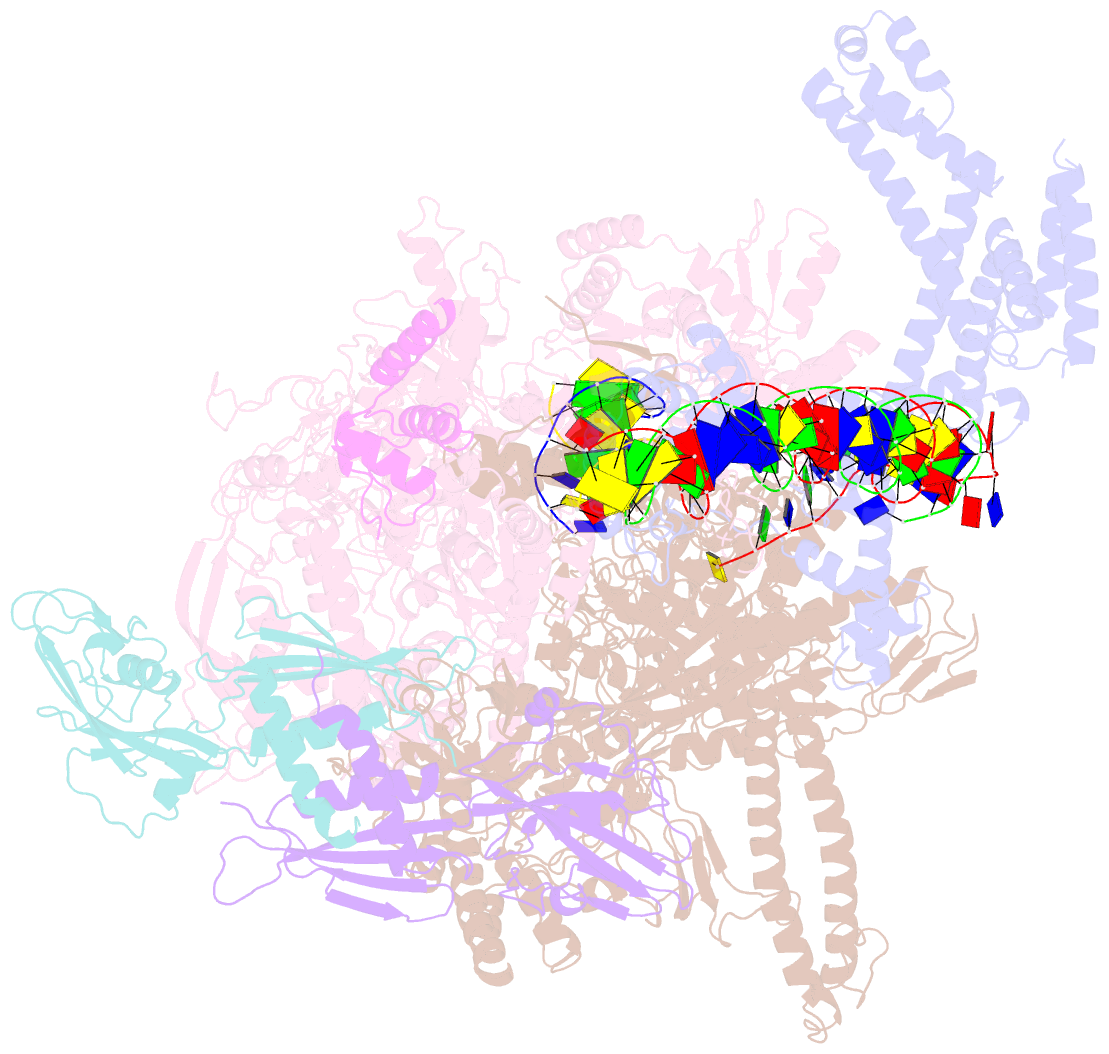
- cryo-EM structure of e. coli rnap sigma70 open complex
- Reference
- Narayanan A, Vago FS, Li K, Qayyum MZ, Yernool D, Jiang W, Murakami KS (2018): "Cryo-EM structure ofEscherichia colisigma70RNA polymerase and promoter DNA complex revealed a role of sigma non-conserved region during the open complex formation." J. Biol. Chem., 293, 7367-7375. doi: 10.1074/jbc.RA118.002161.
- Abstract
- First step of gene expression is transcribing the genetic information stored in DNA to RNA by the transcription machinery including RNA polymerase (RNAP). In Escherichia coli, a primary σ70 factor forms the RNAP holoenzyme to express housekeeping genes. The σ70 contains a large insertion between the conserved regions 1.2 and 2.1, the σ non-conserved region (σNCR), but its function remains to be elucidated. In this study, we determined the cryo-EM structures of the E. coli RNAP σ70 holoenzyme and its complex with promoter DNA (open complex, RPo) at 4.2 and 5.75 Å resolutions, respectively, to reveal native conformations of RNAP and DNA. The RPo structure presented here found an interaction between the σNCR and promoter DNA just upstream of the -10 element, which was not observed in a previously determined E. coli RNAP transcription initiation complex (RPo plus short RNA) structure by X-ray crystallography because of restraint of crystal packing effects. Disruption of the σNCR and DNA interaction by the amino acid substitutions (R157A/R157E) influences the DNA opening around the transcription start site and therefore decreases the transcription activity of RNAP. We propose that the σNCR and DNA interaction is conserved in proteobacteria, and RNAP in other bacteria replaces its role with a transcription factor.